4DJ4
 
 | | X-ray structure of mutant N211D of bifunctional nuclease TBN1 from Solanum lycopersicum (Tomato) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Nuclease, ... | | Authors: | Koval, T, Stepankova, A, Lipovova, P, Podzimek, T, Matousek, J, Duskova, J, Skalova, T, Hasek, J, Dohnalek, J. | | Deposit date: | 2012-02-01 | | Release date: | 2012-11-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Plant multifunctional nuclease TBN1 with unexpected phospholipase activity: structural study and reaction-mechanism analysis.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
6VLX
 
 | | Crystal structure of FabI from Alistipes finegoldii | | Descriptor: | Enoyl-[acyl-carrier-protein] reductase [NADH], GLYCEROL | | Authors: | Radka, C.D, Seetharaman, J, Rock, C.O. | | Deposit date: | 2020-01-27 | | Release date: | 2020-04-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | The genome of a Bacteroidetes inhabitant of the human gut encodes a structurally distinct enoyl-acyl carrier protein reductase (FabI).
J.Biol.Chem., 295, 2020
|
|
8B5R
 
 | | p97-p37-SPI substrate complex | | Descriptor: | I3 sequence being threaded through the p97 channel, Protein phosphatase 1 regulatory subunit 7, Serine/threonine-protein phosphatase PP1-gamma catalytic subunit, ... | | Authors: | van den Boom, J, Marini, G, Meyer, H, Saibil, H. | | Deposit date: | 2022-09-24 | | Release date: | 2023-06-07 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (6.1 Å) | | Cite: | Structural basis of ubiquitin-independent PP1 complex disassembly by p97.
Embo J., 42, 2023
|
|
2FZ5
 
 | | Solution structure of two-electron reduced Megasphaera elsdenii flavodoxin | | Descriptor: | 1-DEOXY-1-(7,8-DIMETHYL-2,4-DIOXO-3,4-DIHYDRO-2H-BENZO[G]PTERIDIN-1-ID-10(5H)-YL)-5-O-PHOSPHONATO-D-RIBITOL, Flavodoxin | | Authors: | van Mierlo, C.P.M, Lijnzaad, P, Vervoort, J, Mueller, F, Berendsen, H.J, de Vlieg, J. | | Deposit date: | 2006-02-09 | | Release date: | 2006-03-14 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Tertiary structure of two-electron reduced Megasphaera elsdenii flavodoxin and some implications, as determined by two-dimensional 1H NMR and restrained molecular dynamics
Eur.J.Biochem., 194, 1990
|
|
4TL9
 
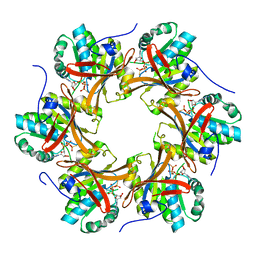 | | Crystal structure of N-terminal C1 domain of KaiC | | Descriptor: | CHLORIDE ION, Circadian clock protein kinase KaiC, MAGNESIUM ION, ... | | Authors: | Abe, J, Hiyama, T.B, Mukaiyama, A, Son, S, Akiyama, S. | | Deposit date: | 2014-05-29 | | Release date: | 2015-07-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.822 Å) | | Cite: | Circadian rhythms. Atomic-scale origins of slowness in the cyanobacterial circadian clock.
Science, 349, 2015
|
|
7A54
 
 | |
6HP0
 
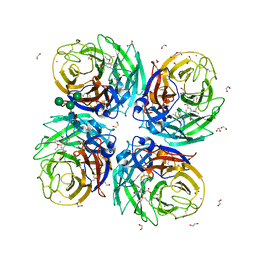 | | Complex of Neuraminidase from H1N1 Influenza Virus in Complex with Oseltamivir Triazol Derivative | | Descriptor: | (3~{R},4~{R},5~{S})-4-acetamido-5-[4-(hydroxymethyl)-1,2,3-triazol-1-yl]-3-pentan-3-yloxy-cyclohexene-1-carboxylic acid, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Pachl, P, Pokorna, J. | | Deposit date: | 2018-09-19 | | Release date: | 2019-09-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Investigation of flexibility of neuraminidase 150-loop using tamiflu derivatives in influenza A viruses H1N1 and H5N1.
Bioorg.Med.Chem., 27, 2019
|
|
6FQN
 
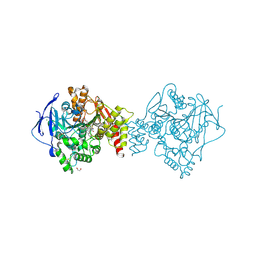 | | Carbamylated T. californica acetylcholineterase bound to uncharged hybrid reactivator 2 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 6-[4-[(7-chloranyl-1,2,3,4-tetrahydroacridin-9-yl)amino]butyl]-2-[(oxidanylamino)methyl]pyridin-3-ol, ... | | Authors: | De la Mora, E, Santoni, G, de Souza, J, Sussman, J, Silman, I, Baati, R, Weik, M, Nachon, F. | | Deposit date: | 2018-02-14 | | Release date: | 2018-08-29 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.30002666 Å) | | Cite: | Structure-Based Optimization of Nonquaternary Reactivators of Acetylcholinesterase Inhibited by Organophosphorus Nerve Agents.
J. Med. Chem., 61, 2018
|
|
7SG0
 
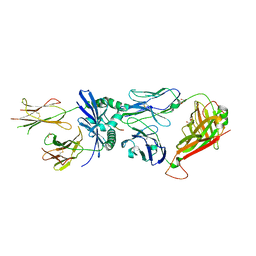 | | W316 TCR in complex with HLA-DQ2-omega1 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, DQ2-glia-omega1 peptide, ... | | Authors: | Ciacchi, L, Farenc, C, Petersen, J, Reid, H.H, Rossjohn, J. | | Deposit date: | 2021-10-04 | | Release date: | 2022-02-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis of T cell receptor specificity and cross-reactivity of two HLA-DQ2.5-restricted gluten epitopes in celiac disease.
J.Biol.Chem., 298, 2022
|
|
7SG1
 
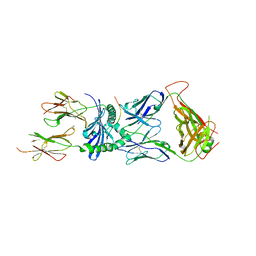 | | XPA5 TCR in complex with HLA-DQ2-alpha1 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Ciacchi, L, Farenc, C, Petersen, J, Reid, H.H, Rossjohn, J. | | Deposit date: | 2021-10-04 | | Release date: | 2022-02-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis of T cell receptor specificity and cross-reactivity of two HLA-DQ2.5-restricted gluten epitopes in celiac disease.
J.Biol.Chem., 298, 2022
|
|
6VLY
 
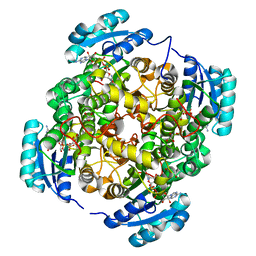 | | Crystal structure of FabI-NADH complex from Alistipes finegoldii | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, CHLORIDE ION, Enoyl-[acyl-carrier-protein] reductase [NADH], ... | | Authors: | Radka, C.D, Seetharaman, J, Rock, C.O. | | Deposit date: | 2020-01-27 | | Release date: | 2020-04-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | The genome of a Bacteroidetes inhabitant of the human gut encodes a structurally distinct enoyl-acyl carrier protein reductase (FabI).
J.Biol.Chem., 295, 2020
|
|
6VVY
 
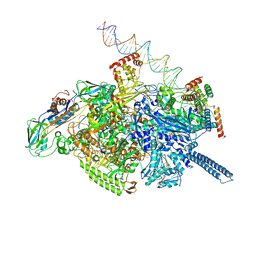 | | Mycobacterium tuberculosis WT RNAP transcription open promoter complex with Sorangicin | | Descriptor: | DNA (65-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Lilic, M, Boyaci, H, Chen, J, Darst, S.A, Campbell, E.A. | | Deposit date: | 2020-02-18 | | Release date: | 2020-10-21 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.42 Å) | | Cite: | The antibiotic sorangicin A inhibits promoter DNA unwinding in a Mycobacterium tuberculosis rifampicin-resistant RNA polymerase.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7SG2
 
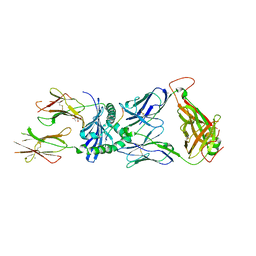 | | XPA5 TCR in complex with HLA-DQ2-omega1 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | Authors: | Ciacchi, L, Farenc, C, Petersen, J, Reid, H.H, Rossjohn, J. | | Deposit date: | 2021-10-04 | | Release date: | 2022-02-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis of T cell receptor specificity and cross-reactivity of two HLA-DQ2.5-restricted gluten epitopes in celiac disease.
J.Biol.Chem., 298, 2022
|
|
1UWI
 
 | |
4S35
 
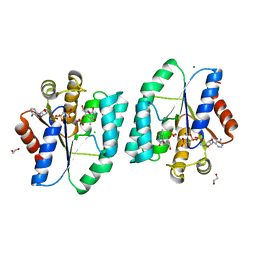 | | AMPPCP and TMP bound Crystal structure of thymidylate kinase (aq_969) from Aquifex Aeolicus VF5 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Biswas, A, Jeyakanthan, J, Sekar, K, Kuramitsu, S, Yokoyama, S. | | Deposit date: | 2015-01-26 | | Release date: | 2016-01-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural studies of a hyperthermophilic thymidylate kinase enzyme reveal conformational substates along the reaction coordinate.
Febs J., 284, 2017
|
|
7SKQ
 
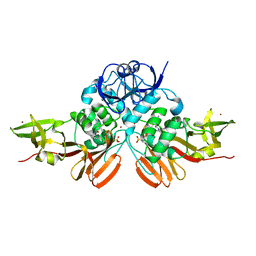 | | BtSCoV-Rf1.2004 Papain-Like protease bound to the non-covalent inhibitor GRL-0617 | | Descriptor: | 3C-like proteinase, 5-amino-2-methyl-N-[(1R)-1-naphthalen-1-ylethyl]benzamide, ZINC ION | | Authors: | Freitas, B, Durie, I, Shepard, J, O'Boyle, B, Enos, S, Pegan, S.D. | | Deposit date: | 2021-10-21 | | Release date: | 2022-03-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.16 Å) | | Cite: | Exploring Noncovalent Protease Inhibitors for the Treatment of Severe Acute Respiratory Syndrome and Severe Acute Respiratory Syndrome-Like Coronaviruses.
Acs Infect Dis., 8, 2022
|
|
7SKR
 
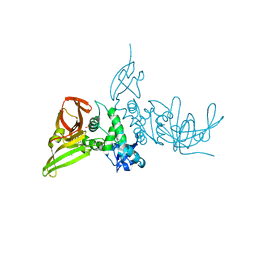 | | BtSCoV-Rf1.2004 Papain-Like protease bound to the non-covalent inhibitor 37 | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, N-[(2-methoxypyridin-4-yl)methyl]-2-[(1R)-1-(naphthalen-1-yl)ethyl]-2-azaspiro[3.3]heptane-6-carboxamide, ... | | Authors: | Durie, I, Shepard, J, Freitas, B, O'Boyle, B, Enos, S, Pegan, S.D. | | Deposit date: | 2021-10-21 | | Release date: | 2022-03-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Exploring Noncovalent Protease Inhibitors for the Treatment of Severe Acute Respiratory Syndrome and Severe Acute Respiratory Syndrome-Like Coronaviruses.
Acs Infect Dis., 8, 2022
|
|
6ZOS
 
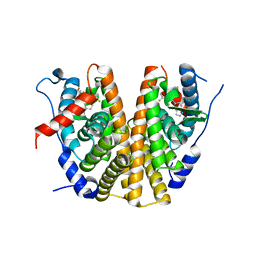 | | Oestrogen receptor ligand binding domain in complex with compound 18 | | Descriptor: | 6-[(6~{S},8~{R})-7-[(1-fluoranylcyclopropyl)methyl]-8-methyl-2,6,8,9-tetrahydropyrazolo[4,3-f]isoquinolin-6-yl]-~{N}-[1-(3-fluoranylpropyl)azetidin-3-yl]pyridin-3-amine, Estrogen receptor | | Authors: | Breed, J. | | Deposit date: | 2020-07-07 | | Release date: | 2021-01-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of AZD9833, a Potent and Orally Bioavailable Selective Estrogen Receptor Degrader and Antagonist.
J.Med.Chem., 63, 2020
|
|
7ZF1
 
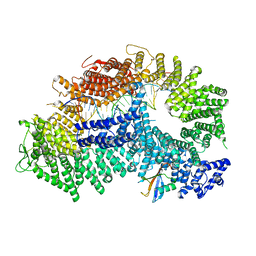 | | Structure of ubiquitinated FANCI in complex with FANCD2 and double-stranded DNA | | Descriptor: | DNA (61-MER), Fanconi anemia group D2 protein, Fanconi anemia group I protein, ... | | Authors: | Lemonidis, K, Rennie, M.L, Arkinson, C, Streetley, J, Clarke, M, Chaugule, V.K, Walden, H. | | Deposit date: | 2022-03-31 | | Release date: | 2022-11-16 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (4.14 Å) | | Cite: | Structural and biochemical basis of interdependent FANCI-FANCD2 ubiquitination.
Embo J., 42, 2023
|
|
3HW1
 
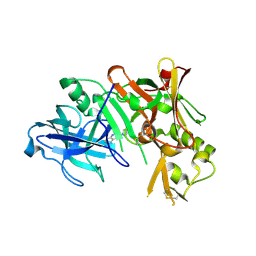 | | Structure of Bace (beta secretase) in complex with ligand EV2 | | Descriptor: | 3-pyrrolidin-1-ylquinoxalin-2-amine, Beta-secretase 1, GLYCEROL | | Authors: | Godemann, R, Madden, J, Kramer, J, Smith, M.A, Barker, J, Ebneth, A. | | Deposit date: | 2009-06-17 | | Release date: | 2009-11-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Fragment-Based Discovery of BACE1 Inhibitors Using Functional Assays
Biochemistry, 48, 2009
|
|
7QDH
 
 | | SARS-CoV-2 S protein S:D614G mutant 1-up | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,Fibritin | | Authors: | Ginex, T, Marco-Marin, C, Wieczor, M, Mata, C.P, Krieger, J, Lopez-Redondo, M.L, Frances-Gomez, C, Ruiz-Rodriguez, P, Melero, R, Sanchez-Sorzano, C.O, Martinez, M, Gougeard, N, Forcada-Nadal, A, Zamora-Caballero, S, Gozalbo-Rovira, R, Sanz-Frasquet, C, Bravo, J, Rubio, V, Marina, A, Geller, R, Comas, I, Gil, C, Coscolla, M, Orozco, M, LLacer, J.L, Carazo, J.M. | | Deposit date: | 2021-11-27 | | Release date: | 2022-05-25 | | Last modified: | 2022-08-10 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | The structural role of SARS-CoV-2 genetic background in the emergence and success of spike mutations: The case of the spike A222V mutation.
Plos Pathog., 18, 2022
|
|
8R8K
 
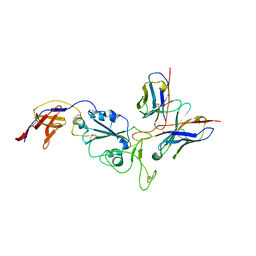 | | XBB-4 Fab in complex with SARS-CoV-2 BA.2.12.1 Spike Glycoprotein | | Descriptor: | Spike glycoprotein,Fibritin, XBB-4 Fab Heavy chain, XBB-4 Fab Light chain | | Authors: | Duyvesteyn, H.M.E, Ren, J, Stuart, D.I. | | Deposit date: | 2023-11-29 | | Release date: | 2024-05-08 | | Last modified: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | A structure-function analysis shows SARS-CoV-2 BA.2.86 balances antibody escape and ACE2 affinity.
Cell Rep Med, 5, 2024
|
|
2C47
 
 | | Structure of casein kinase 1 gamma 2 | | Descriptor: | (2R,3R,4S,5R)-2-(4-AMINO-5-IODO-7H-PYRROLO[2,3-D]PYRIMIDIN-7-YL)-5-(HYDROXYMETHYL)TETRAHYDROFURAN-3,4-DIOL, CASEIN KINASE 1 GAMMA 2 ISOFORM, MAGNESIUM ION | | Authors: | Bunkoczi, G, Rellos, P, Das, S, Ugochukwu, E, Fedorov, O, Sobott, F, Eswaran, J, Amos, A, Ball, L, von Delft, F, Bullock, A, Debreczeni, J, Turnbull, A, Sundstrom, M, Weigelt, J, Arrowsmith, C, Edwards, A, Knapp, S. | | Deposit date: | 2005-10-16 | | Release date: | 2005-11-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Structure of Casein Kinase 1 Gamma 2
To be Published
|
|
8R80
 
 | | SARS-CoV-2 Delta RBD in complex with XBB-9 Fab and an anti-Fab nanobody | | Descriptor: | Spike protein S1, XBB-9 Fab heavy chain, XBB-9 Fab light chain, ... | | Authors: | Zhou, D, Ren, J, Stuart, D.I. | | Deposit date: | 2023-11-27 | | Release date: | 2024-05-08 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (4.03 Å) | | Cite: | A structure-function analysis shows SARS-CoV-2 BA.2.86 balances antibody escape and ACE2 affinity.
Cell Rep Med, 5, 2024
|
|
6FLD
 
 | | Carbamylated T. californica acetylcholineterase bound to uncharged hybrid reactivator 1 | | Descriptor: | 1,2-ETHANEDIOL, 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, 2-[(~{E})-hydroxyiminomethyl]-6-[4-(1,2,3,4-tetrahydroacridin-9-ylamino)butyl]pyridin-3-ol, ... | | Authors: | De la Mora, E, Santoni, G, de Souza, J, Sussman, J, Silman, I, Baati, R, Weik, M, Nachon, F. | | Deposit date: | 2018-01-25 | | Release date: | 2018-08-29 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-Based Optimization of Nonquaternary Reactivators of Acetylcholinesterase Inhibited by Organophosphorus Nerve Agents.
J. Med. Chem., 61, 2018
|
|
