2LTH
 
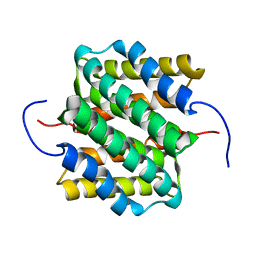 | | NMR structure of major ampullate spidroin 1 N-terminal domain at pH 5.5 | | Descriptor: | Major ampullate spidroin 1 | | Authors: | Otikovs, M, Jaudzems, K, Nordling, K, Landreh, M, Rising, A, Askarieh, G, Knight, S, Johansson, J. | | Deposit date: | 2012-05-25 | | Release date: | 2013-11-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Sequential pH-driven dimerization and stabilization of the N-terminal domain enables rapid spider silk formation.
Nat Commun, 5, 2014
|
|
7XW9
 
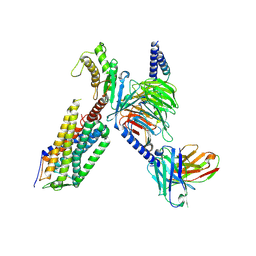 | | Cryo-EM structure of the TRH-bound human TRHR-Gq complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(q) subunit alpha, ... | | Authors: | Ji, S, Dong, Y, Chen, L, Zang, S, Shen, D, Guo, J, Qin, J, Zhang, H, Wang, W, Shen, Q, Mao, C, Zhang, Y. | | Deposit date: | 2022-05-26 | | Release date: | 2022-12-28 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Molecular basis for the activation of thyrotropin-releasing hormone receptor.
Cell Discov, 8, 2022
|
|
2L2O
 
 | |
2L38
 
 | | R29Q Sticholysin II mutant | | Descriptor: | Sticholysin-2 | | Authors: | Castrillo, I, Alegre-Cebollada, J, Martinez-del-Pozo, A, Gavilanes, J, Bruix, M. | | Deposit date: | 2010-09-10 | | Release date: | 2010-09-22 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of StnIIR29Q, a defective lipid binding mutant of the sea anemone actinoporin Sticholysin II
To be Published
|
|
5H3X
 
 | |
2L7U
 
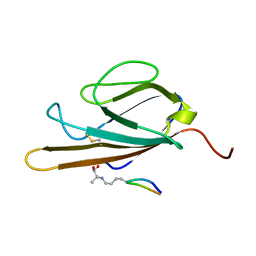 | | Structure of CEL-PEP-RAGE V domain complex | | Descriptor: | Advanced glycosylation end product-specific receptor, Serum albumin peptide | | Authors: | Xue, J, Rai, V, Schmidt, A, Frolov, S, Reverdatto, S, Singer, D, Chabierski, S, Xie, J, Burz, D, Shekhtman, A, Hoffman, R. | | Deposit date: | 2010-12-23 | | Release date: | 2011-05-18 | | Last modified: | 2023-11-29 | | Method: | SOLUTION NMR | | Cite: | Advanced glycation end product recognition by the receptor for AGEs.
Structure, 19, 2011
|
|
4GF0
 
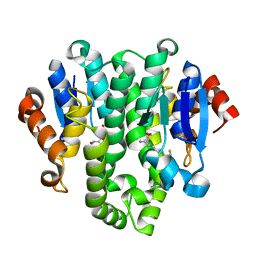 | | Crystal structure of glutahtione transferase homolog from sulfitobacter, TARGET EFI-501084, with bound glutathione | | Descriptor: | CHLORIDE ION, GLUTATHIONE, Glutathione S-transferase | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Armstrong, R.N, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-08-02 | | Release date: | 2012-08-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of glutahtione transferase homolog from sulfitobacter, TARGET EFI-501084, with bound glutathione
To be Published
|
|
4G9H
 
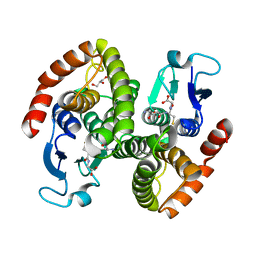 | | Crystal structure of glutahtione s-transferase homolog from yersinia pestis, target EFI-501894, with bound glutathione | | Descriptor: | GLUTATHIONE, GLYCEROL, Glutathione S-transferase | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Armstrong, R.N, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-07-23 | | Release date: | 2012-08-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of glutahtione s-transferase homolog from yersinia pestis, target efi-501894, with bound glutathione
To be Published
|
|
5H1D
 
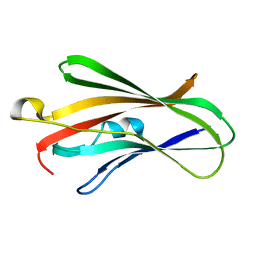 | | Crystal structure of C-terminal of RhoGDI2 | | Descriptor: | Rho GDP-dissociation inhibitor 2 | | Authors: | Liu, J. | | Deposit date: | 2016-10-08 | | Release date: | 2016-10-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.494 Å) | | Cite: | NMR characterization of weak interactions between RhoGDI2 and fragment screening hits.
Biochim. Biophys. Acta, 1861, 2017
|
|
2LRN
 
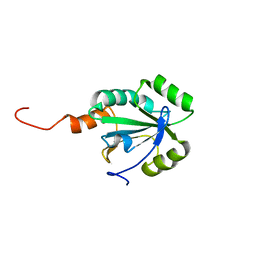 | | Solution structure of a thiol:disulfide interchange protein from Bacteroides sp. | | Descriptor: | Thiol:disulfide interchange protein | | Authors: | Harris, R, Bandaranayake, A.D, Banu, R, Bonanno, J.B, Calarese, D.A, Celikgil, A, Chamala, S, Chan, M.K, Chaparro, R, Evans, B, Garforth, S, Gizzi, A, Hillerich, B, Kar, A, Lafleur, J, Lim, S, Love, J, Matikainen, B, Patel, H, Seidel, R.D, Smith, B, Stead, M, Girvin, M.E, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-04-10 | | Release date: | 2012-04-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a thiol:disulfide interchange protein from Bacteroides sp.
To be Published
|
|
2LST
 
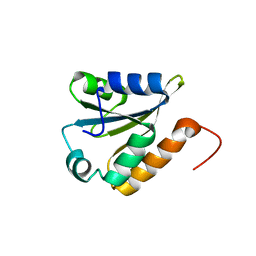 | | Solution structure of a thioredoxin from Thermus thermophilus | | Descriptor: | Thioredoxin | | Authors: | Harris, R, Bandaranayake, A.D, Banu, R, Bonanno, J.B, Calarese, D.A, Celikgil, A, Chamala, S, Chan, M.K, Chaparro, R, Evans, B, Garforth, S, Gizzi, A, Hillerich, B, Kar, A, Lafleur, J, Lim, S, Love, J, Matikainen, B, Patel, H, Seidel, R.D, Smith, B, Stead, M, Girvin, M.E, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-05-04 | | Release date: | 2012-05-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a thioredoxin from Thermus thermophilus
To be Published
|
|
6V0A
 
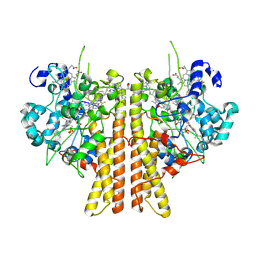 | | Crystal structure of cytochrome c nitrite reductase from the bacterium Geobacter lovleyi with bound sulfate | | Descriptor: | HEME C, Nitrite reductase (cytochrome; ammonia-forming), SULFATE ION | | Authors: | Satyanarayana, L, Campecino, J, Hegg, L.H, Hu, J. | | Deposit date: | 2019-11-18 | | Release date: | 2020-06-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Cytochromecnitrite reductase from the bacteriumGeobacter lovleyirepresents a new NrfA subclass.
J.Biol.Chem., 295, 2020
|
|
2LRT
 
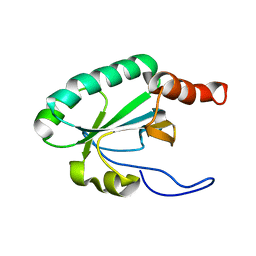 | | Solution structure of the uncharacterized thioredoxin-like protein BVU_1432 from Bacteroides vulgatus | | Descriptor: | Uncharacterized protein | | Authors: | Harris, R, Bandaranayake, A.D, Banu, R, Bonanno, J.B, Calarese, D.A, Celikgil, A, Chamala, S, Chan, M.K, Chaparro, R, Evans, B, Garforth, S, Gizzi, A, Hillerich, B, Kar, A, Lafleur, J, Lim, S, Love, J, Matikainen, B, Patel, H, Seidel, R.D, Smith, B, Stead, M, Girvin, M.E, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-04-13 | | Release date: | 2012-04-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the uncharacterized thioredoxin-like protein BVU_1432 from Bacteroides vulgatus
To be Published
|
|
2LKZ
 
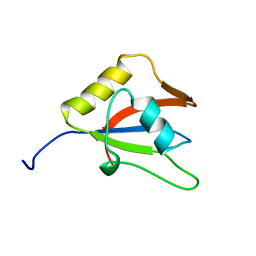 | | Solution structure of the second RRM domain of RBM5 | | Descriptor: | RNA-binding protein 5 | | Authors: | Song, Z, Wu, P, Zhang, J, Wu, J, Shi, Y. | | Deposit date: | 2011-10-23 | | Release date: | 2012-08-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the second RRM domain of RBM5 and its unusual binding characters for different RNA targets
Biochemistry, 2012
|
|
2L8O
 
 | | Solution structure of Chr148 from Cytophaga hutchinsonii, Northeast Structural Genomics Consortium Target Chr148 | | Descriptor: | Activator of Hsp90 ATPase homologue 1-like C-terminal domain-containing protein | | Authors: | Liu, Y, Lee, D, Ciccosanti, C, Nair, L, Rost, B, Acton, T, Xiao, R, Everett, J, Montelione, G, Prestegard, J, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-01-21 | | Release date: | 2011-03-02 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Chr148 from Cytophaga hutchinsonii. Northeast Structural Genomics Consortium Target Chr148
To be Published
|
|
3U93
 
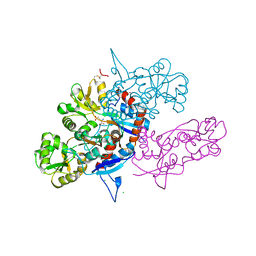 | |
4YBF
 
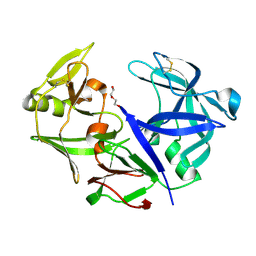 | | Aspartic Proteinase Sapp2 Secreted from Candida Parapsilosis at 1.25 A Resolution | | Descriptor: | Candidapepsin-2, DI(HYDROXYETHYL)ETHER | | Authors: | Dostal, J, Hruskova-Heidingsfeldova, O, Rezacova, P, Brynda, J, Mareckova, L, Pichova, I. | | Deposit date: | 2015-02-18 | | Release date: | 2016-01-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Atomic resolution crystal structure of Sapp2p, a secreted aspartic protease from Candida parapsilosis.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
2XIQ
 
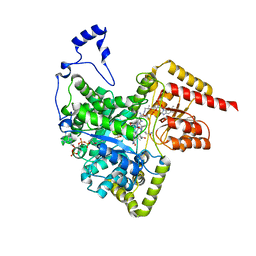 | | Crystal structure of human methylmalonyl-CoA mutase in complex with adenosylcobalamin and malonyl-CoA | | Descriptor: | 5'-DEOXYADENOSINE, COBALAMIN, MALONYL-COENZYME A, ... | | Authors: | Yue, W.W, Froese, D.S, Kochan, G, Chaikuad, A, Krojer, T, Muniz, J, Ugochukwu, E, Arrowsmith, C, Weigelt, J, Edwards, A, Bountra, C, Oppermann, U. | | Deposit date: | 2010-06-30 | | Release date: | 2010-09-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structures of the Human Gtpase Mmaa and Vitamin B12-Dependent Methylmalonyl-Coa Mutase and Insight Into Their Complex Formation.
J.Biol.Chem., 285, 2010
|
|
4Y9T
 
 | | CRYSTAL STRUCTURE OF AN ABC TRANSPORTER SOLUTE BINDING PROTEIN (IPR025997) FROM AGROBACTERIUM VITIS S4 (Avi_5305, TARGET EFI-511224) WITH BOUND ALPHA-D-GLUCOSAMINE | | Descriptor: | 2-amino-2-deoxy-alpha-D-glucopyranose, ABC transporter, solute binding protein | | Authors: | Vetting, M.W, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Koss, J, Wasserman, S.R, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2015-02-17 | | Release date: | 2015-03-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Structure of an ABC transporter solute-binding protein specific for the amino sugars glucosamine and galactosamine.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
4Y7D
 
 | | Alpha/beta hydrolase fold protein from Nakamurella multipartita | | Descriptor: | Alpha/beta hydrolase fold protein, CHLORIDE ION, SODIUM ION | | Authors: | Cuff, M.E, OSIPIUK, J, Holowicki, J, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-02-14 | | Release date: | 2015-02-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Alpha/beta hydrolase fold protein from Nakamurella multipartita.
to be published
|
|
4YPA
 
 | | ASH1L SET domain Q2265A mutant in complex with S-adenosyl methionine (SAM) | | Descriptor: | Histone-lysine N-methyltransferase ASH1L, S-ADENOSYLMETHIONINE, ZINC ION | | Authors: | Rogawski, D.S, Ndoj, J, Cho, H.J, Maillard, I, Grembecka, J, Cierpicki, T. | | Deposit date: | 2015-03-12 | | Release date: | 2015-09-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Two Loops Undergoing Concerted Dynamics Regulate the Activity of the ASH1L Histone Methyltransferase.
Biochemistry, 54, 2015
|
|
4TNH
 
 | | RT XFEL structure of Photosystem II in the dark state at 4.9 A resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Kern, J, Tran, R, Alonso-Mori, R, Koroidov, S, Echols, N, Hattne, J, Ibrahim, M, Gul, S, Laksmono, H, Sierra, R.G, Gildea, R.J, Han, G, Hellmich, J, Lassalle-Kaiser, B, Chatterjee, R, Brewster, A, Stan, C.A, Gloeckner, C, Lampe, A, DiFiore, D, Milathianaki, D, Fry, A.R, Seibert, M.M, Koglin, J.E, Gallo, E, Uhlig, J, Sokaras, D, Weng, T.-C, Zwart, P.H, Skinner, D.E, Bogan, M.J, Messerschmidt, M, Glatzel, P, Williams, G.J, Boutet, S, Adams, P.D, Zouni, A, Messinger, J, Sauter, N.K, Bergmann, U, Yano, J, Yachandra, V.K. | | Deposit date: | 2014-06-04 | | Release date: | 2014-07-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (4.900007 Å) | | Cite: | Taking snapshots of photosynthetic water oxidation using femtosecond X-ray diffraction and spectroscopy.
Nat Commun, 5, 2014
|
|
2XR9
 
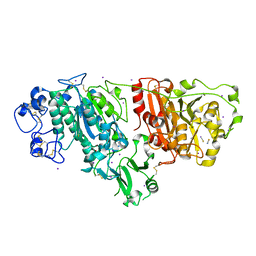 | | Crystal structure of Autotaxin (ENPP2) | | Descriptor: | CALCIUM ION, ECTONUCLEOTIDE PYROPHOSPHATASE/PHOSPHODIESTERASE FAMILY MEMBER 2, IODIDE ION, ... | | Authors: | Kamtekar, S, Hausmann, J, Day, J.E, Christodoulou, E, Perrakis, A. | | Deposit date: | 2010-09-13 | | Release date: | 2011-01-19 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural basis of substrate discrimination and integrin binding by autotaxin.
Nat. Struct. Mol. Biol., 18, 2011
|
|
5X4S
 
 | | Structure of the N-terminal domain (NTD)of SARS-CoV spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Yuan, Y, Zhang, Y, Qi, J, Shi, Y, Gao, G.F. | | Deposit date: | 2017-02-14 | | Release date: | 2017-05-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains
Nat Commun, 8, 2017
|
|
4TNL
 
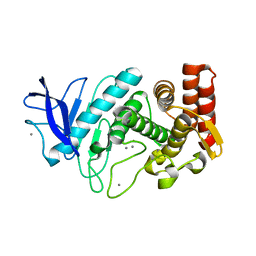 | | 1.8 A resolution room temperature structure of Thermolysin recorded using an XFEL | | Descriptor: | CALCIUM ION, Thermolysin, ZINC ION | | Authors: | Kern, J, Tran, R, Alonso-Mori, R, Koroidov, S, Echols, N, Hattne, J, Ibrahim, M, Gul, S, Laksmono, H, Sierra, R.G, Gildea, R.J, Han, G, Hellmich, J, Lassalle-Kaiser, B, Chatterjee, R, Brewster, A, Stan, C.A, Gloeckner, C, Lampe, A, DiFiore, D, Milathianaki, D, Fry, A.R, Seibert, M.M, Koglin, J.E, Gallo, E, Uhlig, J, Sokaras, D, Weng, T.-C, Zwart, P.H, Skinner, D.E, Bogan, M.J, Messerschmidt, M, Glatzel, P, Williams, G.J, Boutet, S, Adams, P.D, Zouni, A, Messinger, J, Sauter, N.K, Bergmann, U, Yano, J, Yachandra, V.K. | | Deposit date: | 2014-06-04 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Taking snapshots of photosynthetic water oxidation using femtosecond X-ray diffraction and spectroscopy.
Nat Commun, 5, 2014
|
|
