2B5J
 
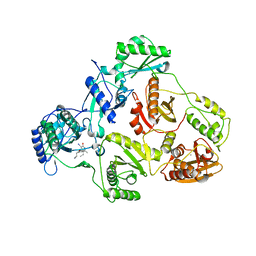 | | Crystal structure of HIV-1 reverse transcriptase (RT) in complex with JANSSEN-R165481 | | Descriptor: | (2E)-3-{3-[(5-ETHYL-3-IODO-6-METHYL-2-OXO-1,2-DIHYDROPYRIDIN-4-YL)OXY]PHENYL}ACRYLONITRILE, MANGANESE (II) ION, Reverse transcriptase P51 SUBUNIT, ... | | Authors: | Himmel, D.H, Das, K, Clark Jr, A.D, Hughes, S.H, Benjahad, A, Oumouch, S, Guillemont, J, Coupa, S, Poncelet, A, Csoka, I, Meyer, C, Andries, K, Mguyen, C.H, Grierson, D.S, Arnold, E. | | Deposit date: | 2005-09-28 | | Release date: | 2005-12-06 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structures for HIV-1 Reverse Transcriptase in Complexes with Three Pyridinone Derivatives: A New Class of Non-Nucleoside Inhibitors Effective against a Broad Range of Drug-Resistant Strains.
J.Med.Chem., 48, 2005
|
|
8ECG
 
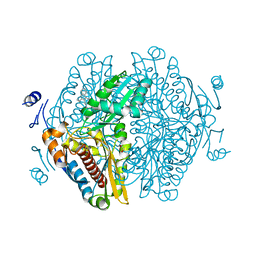 | | Complex of HMG1 with pitavastatin | | Descriptor: | 3-hydroxy-3-methylglutaryl-coenzyme A reductase 1, GLYCEROL, Pitavastatin | | Authors: | Haywood, J, Bond, C.S. | | Deposit date: | 2022-09-01 | | Release date: | 2022-09-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | A fungal tolerance trait and selective inhibitors proffer HMG-CoA reductase as a herbicide mode-of-action.
Nat Commun, 13, 2022
|
|
8DNE
 
 | |
6O0V
 
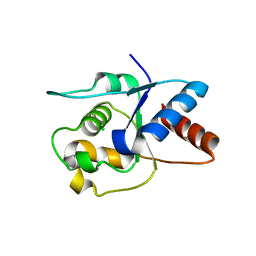 | | Crystal structure of the TIR domain G601P mutant from human SARM1, crystal form 2 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, Sterile alpha and TIR motif-containing protein 1 | | Authors: | Horsefield, S, Burdett, H, Zhang, X, Manik, M.K, Shi, Y, Chen, J, Tiancong, Q, Gilley, J, Lai, J, Gu, W, Rank, M, Casey, L, Ericsson, D.J, Foley, G, Hughes, R.O, Bosanac, T, von Itzstein, M, Rathjen, J.P, Nanson, J.D, Boden, M, Dry, I.B, Williams, S.J, Staskawicz, B.J, Coleman, M.P, Ve, T, Dodds, P.N, Kobe, B. | | Deposit date: | 2019-02-17 | | Release date: | 2019-09-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | NAD+cleavage activity by animal and plant TIR domains in cell death pathways.
Science, 365, 2019
|
|
6YZN
 
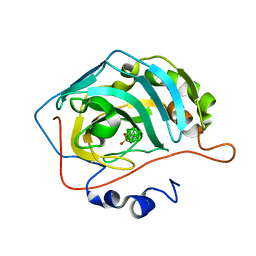 | | Closo-carborane butyl-sulfonamide in complex with CA IX mimic | | Descriptor: | Carbonic anhydrase 2, Carborane closo-butyl-sulfonamide, ZINC ION | | Authors: | Kugler, M, Brynda, J, Pospisilova, K, Rezacova, P. | | Deposit date: | 2020-05-07 | | Release date: | 2020-10-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | The structural basis for the selectivity of sulfonamido dicarbaboranes toward cancer-associated carbonic anhydrase IX.
J Enzyme Inhib Med Chem, 35, 2020
|
|
6YZT
 
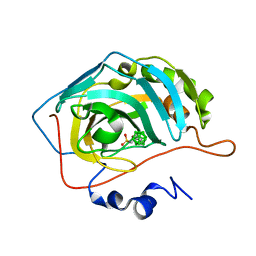 | | Closo-carborane propyl-sulfonamide in complex with CA II | | Descriptor: | Carbonic anhydrase 2, Carborane propyl-sulfonamide, ZINC ION | | Authors: | Kugler, M, Brynda, J, Pospisilova, K, Rezacova, P. | | Deposit date: | 2020-05-07 | | Release date: | 2020-10-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | The structural basis for the selectivity of sulfonamido dicarbaboranes toward cancer-associated carbonic anhydrase IX.
J Enzyme Inhib Med Chem, 35, 2020
|
|
6YU5
 
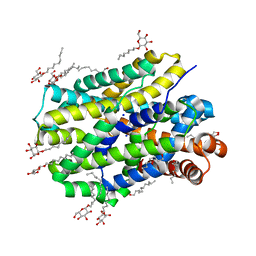 | | Crystal structure of MhsT in complex with L-valine | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, SODIUM ION, Sodium-dependent transporter, ... | | Authors: | Focht, D, Neumann, C, Lyons, J, Eguskiza Bilbao, A, Blunck, R, Malinauskaite, L, Schwarz, I.O, Javitch, J.A, Quick, M, Nissen, P. | | Deposit date: | 2020-04-25 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A non-helical region in transmembrane helix 6 of hydrophobic amino acid transporter MhsT mediates substrate recognition.
Embo J., 40, 2021
|
|
6Z04
 
 | | Nido-carborane butyl-sulfonamide in complex with CA IX mimic | | Descriptor: | Carbonic anhydrase 2, Carborane nido-butyl-sulfonamide, ZINC ION | | Authors: | Kugler, M, Brynda, J, Pospisilova, K, Rezacova, P. | | Deposit date: | 2020-05-07 | | Release date: | 2020-10-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | The structural basis for the selectivity of sulfonamido dicarbaboranes toward cancer-associated carbonic anhydrase IX.
J Enzyme Inhib Med Chem, 35, 2020
|
|
6NMR
 
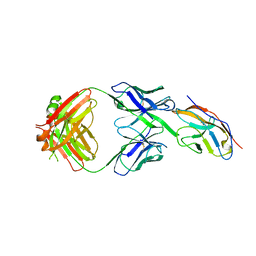 | | Blocking Fab 119 anti-SIRP-alpha antibody in complex with SIRP-alpha Variant 1 | | Descriptor: | Fab 119 anti-SIRP-alpha antibody Variable Heavy Chain, Fab 119 anti-SIRP-alpha antibody Variable Light Chain, Tyrosine-protein phosphatase non-receptor type substrate 1 | | Authors: | Wibowo, A.S, Carter, J.J, Sim, J. | | Deposit date: | 2019-01-11 | | Release date: | 2019-08-07 | | Last modified: | 2019-08-14 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Discovery of high affinity, pan-allelic, and pan-mammalian reactive antibodies against the myeloid checkpoint receptor SIRP alpha.
Mabs, 11, 2019
|
|
5LQP
 
 | |
5LVD
 
 | | Thermolysin in complex with inhibitor (JC67) | | Descriptor: | (2~{S})-4-methyl-2-[[(2~{S})-3-oxidanyl-2-[[oxidanyl(phenylmethoxycarbonylaminomethyl)phosphoryl]amino]propanoyl]amino]pentanoic acid, CALCIUM ION, DIMETHYL SULFOXIDE, ... | | Authors: | Krimmer, S.G, Cramer, J, Heine, A, Klebe, G. | | Deposit date: | 2016-09-14 | | Release date: | 2017-08-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | How Nothing Boosts Affinity: Hydrophobic Ligand Binding to the Virtually Vacated S1' Pocket of Thermolysin.
J. Am. Chem. Soc., 139, 2017
|
|
5ANS
 
 | | Potent and selective inhibitors of MTH1 probe its role in cancer cell survival | | Descriptor: | 1-[4-amino-2-(ethoxymethyl)-1H-imidazo[4,5-c]quinolin-1-yl]-2-methylpropan-2-ol, 7,8-DIHYDRO-8-OXOGUANINE TRIPHOSPHATASE | | Authors: | Kettle, J.G, Alwan, H, Bista, M, Breed, J, Kack, H, Eckersley, K, Foote, K.M, Fillery, S, Goodwin, L, Jones, D, Lau, A, Nissink, J.W.M, Read, J, Scott, J, Taylor, B, Walker, G, Wissler, L. | | Deposit date: | 2015-09-08 | | Release date: | 2016-03-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Potent and Selective Inhibitors of Mth1 Probe its Role in Cancer Cell Survival.
J.Med.Chem., 59, 2016
|
|
6Z4D
 
 | | Crystal Structure of EGFR-T790M/V948R in Complex with Mavelertinib and EAI001 | | Descriptor: | (2R)-2-(1-oxo-1,3-dihydro-2H-isoindol-2-yl)-2-phenyl-N-(1,3-thiazol-2-yl)acetamide, Epidermal growth factor receptor, N-[(3R,4R)-4-fluoro-1-{6-[(3-methoxy-1-methyl-1H-pyrazol-4-yl)amino]-9-methyl-9H-purin-2-yl}pyrrolidin-3-yl]propanamide, ... | | Authors: | Niggenaber, J, Mueller, M.P, Rauh, D. | | Deposit date: | 2020-05-25 | | Release date: | 2020-11-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Complex Crystal Structures of EGFR with Third-Generation Kinase Inhibitors and Simultaneously Bound Allosteric Ligands.
Acs Med.Chem.Lett., 11, 2020
|
|
5ANT
 
 | | Potent and selective inhibitors of MTH1 probe its role in cancer cell survival | | Descriptor: | 2-(2-methoxyethoxy)-6-(methylamino)-9-(phenylmethyl)-7H-purin-8-one, 7,8-DIHYDRO-8-OXOGUANINE TRIPHOSPHATASE | | Authors: | Kettle, J.G, Alwan, H, Bista, M, Breed, J, Kack, H, Eckersley, K, Foote, K.M, Fillery, S, Goodwin, L, Jones, D, Lau, A, Nissink, J.W.M, Read, J, Scott, J, Taylor, B, Walker, G, Wissler, L. | | Deposit date: | 2015-09-08 | | Release date: | 2016-03-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Potent and Selective Inhibitors of Mth1 Probe its Role in Cancer Cell Survival.
J.Med.Chem., 59, 2016
|
|
6V0A
 
 | | Crystal structure of cytochrome c nitrite reductase from the bacterium Geobacter lovleyi with bound sulfate | | Descriptor: | HEME C, Nitrite reductase (cytochrome; ammonia-forming), SULFATE ION | | Authors: | Satyanarayana, L, Campecino, J, Hegg, L.H, Hu, J. | | Deposit date: | 2019-11-18 | | Release date: | 2020-06-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Cytochromecnitrite reductase from the bacteriumGeobacter lovleyirepresents a new NrfA subclass.
J.Biol.Chem., 295, 2020
|
|
5C27
 
 | | Crystal structure of SYK in complex with compound 2 | | Descriptor: | 3-{8-[(3,4-dimethoxyphenyl)amino]imidazo[1,2-a]pyrazin-6-yl}-N-[4-(methylcarbamoyl)phenyl]benzamide, GLU-VAL-TYR-GLU-SER, GLYCEROL, ... | | Authors: | Han, S, Chang, J. | | Deposit date: | 2015-06-15 | | Release date: | 2015-10-07 | | Last modified: | 2016-02-03 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Imidazotriazines: Spleen Tyrosine Kinase (Syk) Inhibitors Identified by Free-Energy Perturbation (FEP).
Chemmedchem, 11, 2016
|
|
8DUA
 
 | | SIV E660.CR54 SOS-2P Env Trimer with ITS92.02 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp120, Envelope glycoprotein gp41, ... | | Authors: | Gorman, J, Kwong, P.D. | | Deposit date: | 2022-07-27 | | Release date: | 2022-10-26 | | Last modified: | 2022-11-30 | | Method: | ELECTRON MICROSCOPY (4.32 Å) | | Cite: | Cryo-EM structures of prefusion SIV envelope trimer.
Nat.Struct.Mol.Biol., 29, 2022
|
|
4OV4
 
 | | Isopropylmalate synthase binding with ketoisovalerate | | Descriptor: | 2-isopropylmalate synthase, 3-METHYL-2-OXOBUTANOIC ACID, ZINC ION | | Authors: | Zhang, Z, Wu, J, Wang, C, Zhang, P. | | Deposit date: | 2014-02-20 | | Release date: | 2014-08-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Subdomain II of alpha-isopropylmalate synthase is essential for activity: inferring a mechanism of feedback inhibition.
J.Biol.Chem., 289, 2014
|
|
2ANY
 
 | | Expression, Crystallization and the Three-dimensional Structure of the Catalytic Domain of Human Plasma Kallikrein: Implications for Structure-Based Design of Protease Inhibitors | | Descriptor: | BENZAMIDINE, PHOSPHATE ION, plasma kallikrein, ... | | Authors: | Tang, J, Yu, C.L, Williams, S.R, Springman, E, Jeffery, D, Sprengeler, P.A, Estevez, A, Sampang, J, Shrader, W, Spencer, J.R, Young, W.B, McGrath, M.E, Katz, B.A. | | Deposit date: | 2005-08-11 | | Release date: | 2005-10-11 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Expression, crystallization, and three-dimensional structure of the catalytic domain of human plasma kallikrein.
J.Biol.Chem., 280, 2005
|
|
6YVD
 
 | | Head segment of the S.cerevisiae condensin holocomplex in presence of ATP | | Descriptor: | Condensin complex subunit 2, Condensin complex subunit 3, Structural maintenance of chromosomes protein 2, ... | | Authors: | Merkel, F, Haering, C.H, Hassler, M, Lee, B.G, Lowe, J. | | Deposit date: | 2020-04-28 | | Release date: | 2020-07-22 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (7.6 Å) | | Cite: | Cryo-EM structures of holo condensin reveal a subunit flip-flop mechanism.
Nat.Struct.Mol.Biol., 27, 2020
|
|
5JWR
 
 | | Crystal structure of foldswitch-stabilized KaiB in complex with the N-terminal CI domain of KaiC and a dimer of KaiA C-terminal domains from Thermosynechococcus elongatus | | Descriptor: | Circadian clock protein KaiA, Circadian clock protein KaiB, Circadian clock protein kinase KaiC, ... | | Authors: | Tseng, R, Goularte, N.F, Chavan, A, Luu, J, Chang, Y.G, Heilser, J, Tripathi, S, LiWang, A, Partch, C.L. | | Deposit date: | 2016-05-12 | | Release date: | 2017-03-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Structural basis of the day-night transition in a bacterial circadian clock.
Science, 355, 2017
|
|
6YZS
 
 | | Carborane closo-pentyl-sulfonamide in complex with CA II | | Descriptor: | Carbonic anhydrase 2, Carborane closo-pentyl-sulfonamide, SODIUM ION, ... | | Authors: | Kugler, M, Brynda, J, Pospisilova, K, Rezacova, P. | | Deposit date: | 2020-05-07 | | Release date: | 2020-10-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | The structural basis for the selectivity of sulfonamido dicarbaboranes toward cancer-associated carbonic anhydrase IX.
J Enzyme Inhib Med Chem, 35, 2020
|
|
5JRP
 
 | | crystal structure of monoclonal antibody MR78 Fab | | Descriptor: | SODIUM ION, marberg virus monoclonal antibody MR78 Fab heavy chain, marberg virus monoclonal antibody MR78 Fab light chain | | Authors: | Dong, J, Crowe, J. | | Deposit date: | 2016-05-06 | | Release date: | 2017-11-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Role of Non-local Interactions between CDR Loops in Binding Affinity of MR78 Antibody to Marburg Virus Glycoprotein.
Structure, 25, 2017
|
|
2X1P
 
 | | Gelsolin Nanobody | | Descriptor: | GELSOLIN NANOBODY | | Authors: | Van Den Abbeele, A, Declercq, S, De Ganck, A, De Corte, V, Van Loo, B, Srinivasan, V, Steyaert, J, Van De Kerckhove, J, Gettemans, J. | | Deposit date: | 2010-01-03 | | Release date: | 2011-01-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | A Llama-Derived Gelsolin Single-Domain Antibody Blocks Gelsolin-G-Actin Interaction.
Cell.Mol.Life Sci., 67, 2010
|
|
2ANW
 
 | | Expression, crystallization and three-dimensional structure of the catalytic domain of human plasma kallikrein: Implications for structure-based design of protease inhibitors | | Descriptor: | BENZAMIDINE, plasma kallikrein, light chain | | Authors: | Tang, J, Yu, C.L, Williams, S.R, Springman, E, Jeffery, D, Sprengeler, P.A, Estevez, A, Sampang, J, Shrader, W, Spencer, J.R, Young, W.B, McGrath, M.E, Katz, B.A. | | Deposit date: | 2005-08-11 | | Release date: | 2005-10-11 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Expression, crystallization, and three-dimensional structure of the catalytic domain of human plasma kallikrein.
J.Biol.Chem., 280, 2005
|
|
