6TTE
 
 | | Beta-galactosidase in complex with PETG | | 分子名称: | 2-phenylethyl 1-thio-beta-D-galactopyranoside, Beta-galactosidase, MAGNESIUM ION | | 著者 | Saur, M, Hartshorn, M.J, Dong, J, Reeks, J, Bunkoczi, G, Jhoti, H, Williams, P.A. | | 登録日 | 2019-12-27 | | 公開日 | 2020-01-15 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (2.2 Å) | | 主引用文献 | Fragment-based drug discovery using cryo-EM.
Drug Discov Today, 25, 2020
|
|
3K3Q
 
 | | Crystal Structure of a Llama Antibody complexed with the C. Botulinum Neurotoxin Serotype A Catalytic Domain | | 分子名称: | Botulinum neurotoxin type A, ZINC ION, llama Aa1 VHH domain | | 著者 | Thompson, A.A, Dong, J, Marks, J.D, Stevens, R.C. | | 登録日 | 2009-10-04 | | 公開日 | 2010-02-23 | | 最終更新日 | 2017-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | A Single-Domain Llama Antibody Potently Inhibits the Enzymatic Activity of Botulinum Neurotoxin by Binding to the Non-Catalytic alpha-Exosite Binding Region.
J.Mol.Biol., 397, 2010
|
|
8J37
 
 | | Crystal structure of SARS-Cov-2 main protease K90R mutant in complex with PF00835231 | | 分子名称: | 3C-like proteinase nsp5, N-[(2S)-1-({(2S,3S)-3,4-dihydroxy-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-4-methoxy-1H-indole-2-carboxamide | | 著者 | Zhou, X.L, Lin, C, Zou, X.F, Zhang, J, Li, J. | | 登録日 | 2023-04-16 | | 公開日 | 2024-04-17 | | 実験手法 | X-RAY DIFFRACTION (1.68 Å) | | 主引用文献 | Crystal structure of SARS-Cov-2 main protease
K90R mutant in complex with PF00835231
To Be Published
|
|
8J35
 
 | | Crystal structure of SARS-Cov-2 main protease G15S mutant in complex with PF00835231 | | 分子名称: | 3C-like proteinase nsp5, N-[(2S)-1-({(2S,3S)-3,4-dihydroxy-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-4-methoxy-1H-indole-2-carboxamide | | 著者 | Zhou, X.L, Lin, C, Zou, X.F, Zhang, J, Li, J. | | 登録日 | 2023-04-16 | | 公開日 | 2024-04-17 | | 実験手法 | X-RAY DIFFRACTION (1.79 Å) | | 主引用文献 | Crystal structure of SARS-Cov-2 main protease
G15S mutant in complex with PF00835231
To Be Published
|
|
8J3A
 
 | | Crystal structure of SARS-Cov-2 main protease Y54C mutant in complex with PF00835231 | | 分子名称: | 3C-like proteinase nsp5, N-[(2S)-1-({(2S,3S)-3,4-dihydroxy-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-4-methoxy-1H-indole-2-carboxamide | | 著者 | Zhou, X.L, Lin, C, Zou, X.F, Zhang, J, Li, J. | | 登録日 | 2023-04-16 | | 公開日 | 2024-04-17 | | 実験手法 | X-RAY DIFFRACTION (1.91 Å) | | 主引用文献 | Crystal structure of SARS-Cov-2 main protease
Y54C mutant in complex with PF00835231
To Be Published
|
|
3BUX
 
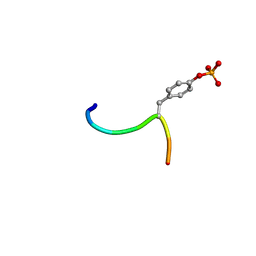 | | Crystal structure of c-Cbl-TKB domain complexed with its binding motif in c-Met | | 分子名称: | 13-meric peptide from Hepatocyte growth factor receptor, E3 ubiquitin-protein ligase CBL | | 著者 | Ng, C, Jackson, R.A, Buschdorf, J.P, Sun, Q, Guy, G.R, Sivaraman, J. | | 登録日 | 2008-01-03 | | 公開日 | 2008-02-26 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.35 Å) | | 主引用文献 | Structural basis for a novel intrapeptidyl H-bond and reverse binding of c-Cbl-TKB domain substrates
Embo J., 27, 2008
|
|
8RBS
 
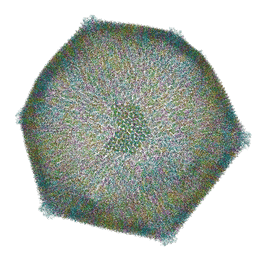 | | Emiliania huxleyi virus 201 (EhV-201) asymmetrical unit of capsid proteins predicted by AlphaFold2 fitted into the cryo-EM density of EhV-201 virion composite map. | | 分子名称: | Major capsid protein, Penton protein | | 著者 | Homola, M, Buttner, C.R, Fuzik, T, Novacek, J, Chaillet, M, Forster, F, Plevka, P. | | 登録日 | 2023-12-04 | | 公開日 | 2023-12-20 | | 最終更新日 | 2024-04-24 | | 実験手法 | ELECTRON MICROSCOPY (18 Å) | | 主引用文献 | Structure and replication cycle of a virus infecting climate-modulating alga Emiliania huxleyi.
Sci Adv, 10, 2024
|
|
8RBT
 
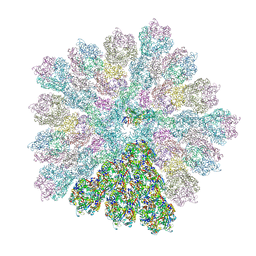 | | Emiliania huxleyi virus 201 (EhV-201) capsid proteins predicted by AlphaFold2 fitted into a cryo-EM density map of the EhV-201 virion capsid. | | 分子名称: | Major capsid protein, Penton protein | | 著者 | Homola, M, Buttner, C.R, Fuzik, T, Novacek, J, Chaillet, M, Forster, F, Plevka, P. | | 登録日 | 2023-12-04 | | 公開日 | 2023-12-20 | | 最終更新日 | 2024-04-24 | | 実験手法 | ELECTRON MICROSCOPY (12 Å) | | 主引用文献 | Structure and replication cycle of a virus infecting climate-modulating alga Emiliania huxleyi.
Sci Adv, 10, 2024
|
|
6NQ0
 
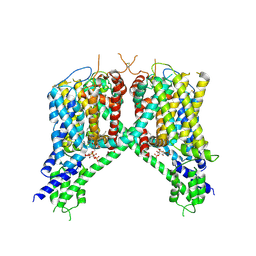 | | Cryo-EM structure of human TPC2 channel in the ligand-bound open state | | 分子名称: | (2R)-3-{[(S)-hydroxy{[(1S,2R,3R,4S,5S,6R)-2,4,6-trihydroxy-3,5-bis(phosphonooxy)cyclohexyl]oxy}phosphoryl]oxy}propane-1,2-diyl dioctanoate, Two pore calcium channel protein 2 | | 著者 | She, J, Zeng, W, Guo, J, Chen, Q, Bai, X, Jiang, Y. | | 登録日 | 2019-01-18 | | 公開日 | 2019-03-27 | | 最終更新日 | 2019-11-20 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Structural mechanisms of phospholipid activation of the human TPC2 channel.
Elife, 8, 2019
|
|
3BXM
 
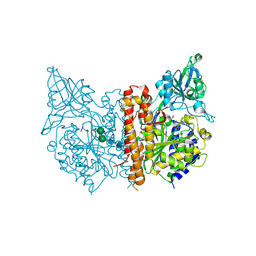 | | Structure of an inactive mutant of human glutamate carboxypeptidase II [GCPII(E424A)] in complex with N-acetyl-Asp-Glu (NAAG) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | 著者 | Lubkowski, J, Barinka, C. | | 登録日 | 2008-01-14 | | 公開日 | 2009-01-27 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.71 Å) | | 主引用文献 | Reaction mechanism of glutamate carboxypeptidase II revealed by mutagenesis, X-ray crystallography, and computational methods.
Biochemistry, 48, 2009
|
|
8J32
 
 | | Crystal structure of SARS-Cov-2 main protease in complex with PF00835231 | | 分子名称: | 3C-like proteinase nsp5, N-[(2S)-1-({(2S,3S)-3,4-dihydroxy-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-4-methoxy-1H-indole-2-carboxamide | | 著者 | Zhou, X.L, Lin, C, Zou, X.F, Zhang, J, Li, J. | | 登録日 | 2023-04-16 | | 公開日 | 2024-04-17 | | 実験手法 | X-RAY DIFFRACTION (2.21 Å) | | 主引用文献 | Crystal structure of SARS-Cov-2 main protease in complex with PF00835231
To Be Published
|
|
8J38
 
 | | Crystal structure of SARS-Cov-2 main protease P132H mutant in complex with PF00835231 | | 分子名称: | 3C-like proteinase nsp5, N-[(2S)-1-({(2S,3S)-3,4-dihydroxy-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-4-methoxy-1H-indole-2-carboxamide | | 著者 | Zhou, X.L, Lin, C, Zou, X.F, Zhang, J, Li, J. | | 登録日 | 2023-04-16 | | 公開日 | 2024-04-17 | | 実験手法 | X-RAY DIFFRACTION (1.72 Å) | | 主引用文献 | Crystal structure of SARS-Cov-2 main protease
P132H mutant in complex with PF00835231
To Be Published
|
|
8J34
 
 | | Crystal structure of MERS main protease in complex with PF00835231 | | 分子名称: | N-[(2S)-1-({(2S,3S)-3,4-dihydroxy-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-4-methoxy-1H-indole-2-carboxamide, ORF1a | | 著者 | Zhou, X.L, Lin, C, Zou, X.F, Zhang, J, Li, J. | | 登録日 | 2023-04-16 | | 公開日 | 2024-04-17 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystal structure of MERS main protease in complex with PF00835231
To Be Published
|
|
8J3B
 
 | | Crystal structure of SARS-Cov-2 main protease S46F mutant in complex with PF00835231 | | 分子名称: | 3C-like proteinase nsp5, N-[(2S)-1-({(2S,3S)-3,4-dihydroxy-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-4-methoxy-1H-indole-2-carboxamide | | 著者 | Zhou, X.L, Lin, C, Zou, X.F, Zhang, J, Li, J. | | 登録日 | 2023-04-16 | | 公開日 | 2024-04-17 | | 実験手法 | X-RAY DIFFRACTION (1.64 Å) | | 主引用文献 | Crystal structure of SARS-Cov-2 main protease
S46F mutant in complex with PF00835231
To Be Published
|
|
3BYV
 
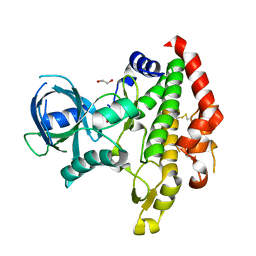 | | Crystal structure of Toxoplasma gondii specific rhoptry antigen kinase domain | | 分子名称: | 1,2-ETHANEDIOL, MAGNESIUM ION, Rhoptry kinase | | 著者 | Wernimont, A.K, Lunin, V.V, Yang, C, Lew, J, Kozieradzki, I, Lin, Y.H, Sun, X, Khuu, C, Zhao, Y, Schapira, M, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bochkarev, A, Hui, R, Sibley, D, Qiu, W, Structural Genomics Consortium (SGC) | | 登録日 | 2008-01-16 | | 公開日 | 2008-01-29 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Novel structural and regulatory features of rhoptry secretory kinases in Toxoplasma gondii.
Embo J., 28, 2009
|
|
5A72
 
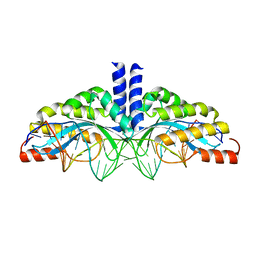 | | Crystal structure of the homing endonuclease I-CvuI in complex with its target (Sro1.3) in the presence of 2 mM Ca | | 分子名称: | 24MER DNA, 5'-D(*DTP*CP*AP*GP*AP*AP*CP*GP*TP*CP*GP*TP*AP *DCP*GP*AP*CP*GP*TP*TP*CP*TP*GP*A)-3', CALCIUM ION, ... | | 著者 | Molina, R, Redondo, P, LopezMendez, B, Villate, M, Merino, N, Blanco, F.J, Valton, J, Grizot, S, Duchateau, P, Prieto, J, Montoya, G. | | 登録日 | 2015-07-02 | | 公開日 | 2015-09-23 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Crystal Structure of the Homing Endonuclease I-Cvui Provides a New Template for Genome Modification
J.Biol.Chem., 290, 2015
|
|
8J39
 
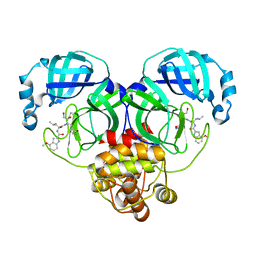 | | Crystal structure of SARS-Cov-2 main protease V186F mutant in complex with PF00835231 | | 分子名称: | 3C-like proteinase nsp5, N-[(2S)-1-({(2S,3S)-3,4-dihydroxy-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-4-methoxy-1H-indole-2-carboxamide | | 著者 | Zhou, X.L, Lin, C, Zou, X.F, Zhang, J, Li, J. | | 登録日 | 2023-04-16 | | 公開日 | 2024-04-17 | | 実験手法 | X-RAY DIFFRACTION (1.66 Å) | | 主引用文献 | Crystal structure of SARS-Cov-2 main protease
V186F mutant in complex with PF00835231
To Be Published
|
|
6H4B
 
 | | A polyamorous repressor: deciphering the evolutionary strategy used by the phage-inducible chromosomal islands to spread in nature. | | 分子名称: | ORF026, Orf20 | | 著者 | Ciges-Tomas, J.R, Alite, C, Bowring, J.Z, Donderis, J, Penades, J.R, Marina, A. | | 登録日 | 2018-07-20 | | 公開日 | 2019-08-28 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.902 Å) | | 主引用文献 | The structure of a polygamous repressor reveals how phage-inducible chromosomal islands spread in nature.
Nat Commun, 10, 2019
|
|
6TSK
 
 | | Beta-galactosidase in complex with L-ribose | | 分子名称: | Beta-galactosidase, MAGNESIUM ION, beta-L-ribopyranose | | 著者 | Saur, M, Hartshorn, M.J, Dong, J, Reeks, J, Bunkoczi, G, Jhoti, H, Williams, P.A. | | 登録日 | 2019-12-20 | | 公開日 | 2020-01-08 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (2.3 Å) | | 主引用文献 | Fragment-based drug discovery using cryo-EM.
Drug Discov Today, 25, 2020
|
|
6H5B
 
 | |
8J36
 
 | | Crystal structure of SARS-Cov-2 main protease M49I mutant in complex with PF00835231 | | 分子名称: | 3C-like proteinase nsp5, N-[(2S)-1-({(2S,3S)-3,4-dihydroxy-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-4-methoxy-1H-indole-2-carboxamide | | 著者 | Zhou, X.L, Lin, C, Zou, X.F, Zhang, J, Li, J. | | 登録日 | 2023-04-16 | | 公開日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (2.21 Å) | | 主引用文献 | Crystal structure of SARS-Cov-2 main protease
M49I mutant in complex with PF00835231
To Be Published
|
|
3BUM
 
 | | Crystal structure of c-Cbl-TKB domain complexed with its binding motif in Sprouty2 | | 分子名称: | E3 ubiquitin-protein ligase CBL, Protein sprouty homolog 2 | | 著者 | Ng, C, Jackson, A.R, Buschdorf, P.J, Sun, Q, Guy, R.G, Sivaraman, J. | | 登録日 | 2008-01-03 | | 公開日 | 2008-02-26 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural basis for a novel intrapeptidyl H-bond and reverse binding of c-Cbl-TKB domain substrates
Embo J., 27, 2008
|
|
3BUW
 
 | | Crystal structure of c-Cbl-TKB domain complexed with its binding motif in Syk | | 分子名称: | 13-meric peptide from Tyrosine-protein kinase SYK, E3 ubiquitin-protein ligase CBL | | 著者 | Ng, C, Jackson, R.A, Buschdorf, J.P, Sun, Q, Guy, G.R, Sivaraman, J. | | 登録日 | 2008-01-03 | | 公開日 | 2008-02-26 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Structural basis for a novel intrapeptidyl H-bond and reverse binding of c-Cbl-TKB domain substrates
Embo J., 27, 2008
|
|
4ET9
 
 | | Hen egg-white lysozyme solved from 5 fs free-electron laser pulse data | | 分子名称: | CHLORIDE ION, Lysozyme C, SODIUM ION | | 著者 | Boutet, S, Lomb, L, Williams, G, Barends, T, Aquila, A, Doak, R.B, Weierstall, U, DePonte, D, Steinbrener, J, Shoeman, R, Messerschmidt, M, Barty, A, White, T, Kassemeyer, S, Kirian, R, Seibert, M, Montanez, P, Kenney, C, Herbst, R, Hart, P, Pines, J, Haller, G, Gruner, S, Philllip, H, Tate, M, Hromalik, M, Koerner, L, van Bakel, N, Morse, J, Ghonsalves, W, Arnlund, D, Bogan, M, Calemann, C, Fromme, R, Hampton, C, Hunter, M, Johansson, L, Katona, G, Kupitz, C, Liang, M, Martin, A, Nass, K, Redecke, L, Stellato, F, Timneanu, N, Wang, D, Zatsepin, N, Schafer, D, Defever, K, Neutze, R, Fromme, P, Spence, J, Chapman, H, Schlichting, I. | | 登録日 | 2012-04-24 | | 公開日 | 2012-06-13 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | High-resolution protein structure determination by serial femtosecond crystallography.
Science, 337, 2012
|
|
4ETD
 
 | | Lysozyme, room-temperature, rotating anode, 0.0026 MGy | | 分子名称: | CHLORIDE ION, Lysozyme C | | 著者 | Boutet, S, Lomb, L, Williams, G, Barends, T, Aquila, A, Doak, R.B, Weierstall, U, DePonte, D, Steinbrener, J, Shoeman, R, Messerschmidt, M, Barty, A, White, T, Kassemeyer, S, Kirian, R, Seibert, M, Montanez, P, Kenney, C, Herbst, R, Hart, P, Pines, J, Haller, G, Gruner, S, Philllip, H, Tate, M, Hromalik, M, Koerner, L, van Bakel, N, Morse, J, Ghonsalves, W, Arnlund, D, Bogan, M, Calemann, C, Fromme, R, Hampton, C, Hunter, M, Johansson, L, Katona, G, Kupitz, C, Liang, M, Martin, A, Nass, K, Redecke, L, Stellato, F, Timneanu, N, Wang, D, Zatsepin, N, Schafer, D, Defever, K, Neutze, R, Fromme, P, Spence, J, Chapman, H, Schlichting, I. | | 登録日 | 2012-04-24 | | 公開日 | 2012-06-13 | | 最終更新日 | 2017-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.904 Å) | | 主引用文献 | High-resolution protein structure determination by serial femtosecond crystallography.
Science, 337, 2012
|
|
