4AFL
 
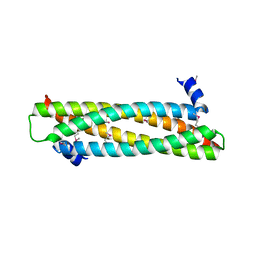 | | The crystal structure of the ING4 dimerization domain reveals the functional organization of the ING family of chromatin binding proteins. | | Descriptor: | INHIBITOR OF GROWTH PROTEIN 4 | | Authors: | Culurgioni, S, Munoz, I.G, Moreno, A, Palacios, A, Villate, M, Palmero, I, Montoya, G, Blanco, F.J. | | Deposit date: | 2012-01-19 | | Release date: | 2012-02-22 | | Last modified: | 2012-07-11 | | Method: | X-RAY DIFFRACTION (2.275 Å) | | Cite: | Crystal Structure of Inhibitor of Growth 4 (Ing4) Dimerization Domain Reveals Functional Organization of Ing Family of Chromatin-Binding Proteins.
J.Biol.Chem., 287, 2012
|
|
4D2G
 
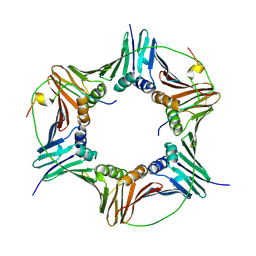 | | Crystal structure of human PCNA in complex with p15 peptide | | Descriptor: | P15, PROLIFERATING CELL NUCLEAR ANTIGEN | | Authors: | DeBiasio, A, Ibanez, A, Mortuza, G, Molina, R, Cordeiro, T.N, Castillo, F, Villate, M, Merino, N, Lelli, M, Diercks, T, Luque, I, Bernardo, P, Montoya, G, Blanco, F.J. | | Deposit date: | 2014-05-09 | | Release date: | 2015-03-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure of P15(Paf)-PCNA Complex and Implications for Clamp Sliding During DNA Replication and Repair.
Nat.Commun., 6, 2015
|
|
2XE0
 
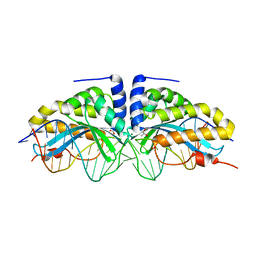 | | Molecular basis of engineered meganuclease targeting of the endogenous human RAG1 locus | | Descriptor: | 24MER DNA, ACETATE ION, I-CREI V2V3 VARIANT, ... | | Authors: | Munoz, I.G, Prieto, J, Subramanian, S, Coloma, J, Redondo, P, Villate, M, Merino, N, Marenchino, M, D'Abramo, M, Gervasio, F.L, Grizot, S, Daboussi, F, Smith, J, Chion-Sotine, I, Paques, F, Duchateau, P, Alibes, A, Stricher, F, Serrano, L, Blanco, F.J, Montoya, G. | | Deposit date: | 2010-05-10 | | Release date: | 2010-09-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Molecular Basis of Engineered Meganuclease Targeting of the Endogenous Human Rag1 Locus
Nucleic Acids Res., 39, 2011
|
|
5A74
 
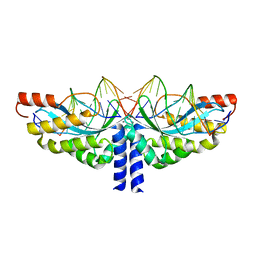 | | Crystal structure of the homing endonuclease I-CvuI in complex with its target (Sro1.3) in the presence of 2 mM Mn | | Descriptor: | 10MER DNA, 5'-D(*GP*AP*CP*GP*TP*TP*CP*TP*GP*AP)-3', 14MER DNA, ... | | Authors: | Molina, R, Redondo, P, LopezMendez, B, Villate, M, Merino, N, Blanco, F.J, Valton, J, Grizot, S, Duchateau, P, Prieto, J, Montoya, G. | | Deposit date: | 2015-07-02 | | Release date: | 2015-09-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of the Homing Endonuclease I-Cvui Provides a New Template for Genome Modification
J.Biol.Chem., 290, 2015
|
|
5A0M
 
 | | THE CRYSTAL STRUCTURE OF I-SCEI IN COMPLEX WITH ITS TARGET DNA IN THE PRESENCE OF MN | | Descriptor: | 5'-D(*CP*AP*CP*GP*CP*TP*AP*GP*GP*GP*AP*TP*AP*AP)-3', 5'-D(*CP*AP*GP*GP*GP*TP*AP*AP*TP*AP*CP)-3', 5'-D(*CP*CP*CP*TP*AP*GP*CP*GP*TP)-3', ... | | Authors: | Prieto, J, Redondo, P, Merino, N, Villate, M, Blanco, F.J, Molina, R. | | Deposit date: | 2015-04-21 | | Release date: | 2016-05-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of the I-Scei Nuclease Complexed with its DsDNA Target and Three Catalytic Metal Ions.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
2XAM
 
 | | Inositol 1,3,4,5,6-pentakisphosphate 2-kinase from A. thaliana in complex with ADP and IP6. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, INOSITOL-PENTAKISPHOSPHATE 2-KINASE, ... | | Authors: | Gonzalez, B, Banos-Sanz, J.I, Villate, M, Brearley, C.A, Sanz-Aparicio, J. | | Deposit date: | 2010-03-31 | | Release date: | 2010-05-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Inositol 1,3,4,5,6-Pentakisphosphate 2-Kinase is a Distant Ipk Member with a Singular Inositide Binding Site for Axial 2-Oh Recognition.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
5A77
 
 | | Crystal structure of the homing endonuclease I-CvuI in complex with I- CreI target (C1221) in the presence of 2 mM Mg revealing DNA cleaved | | Descriptor: | 10MER DNA, 5'-D(*GP*AP*CP*GP*TP*TP*TP*TP* GP*AP*DGP*AP*CP*GP*TP*TP*TP*TP*GP*A)-3', 14MER DNA, ... | | Authors: | Molina, R, Redondo, P, LopezMendez, B, Villate, M, Merino, N, Blanco, F.J, Valton, J, Grizot, S, Duchateau, P, Prieto, J, Montoya, G. | | Deposit date: | 2015-07-03 | | Release date: | 2015-09-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of the Homing Endonuclease I-Cvui Provides a New Template for Genome Modification
J.Biol.Chem., 290, 2015
|
|
5A78
 
 | | Crystal structure of the homing endonuclease I-CvuI in complex with I- CreI target (C1221) in the presence of 2 mM Mg revealing DNA not cleaved | | Descriptor: | 24MER DNA, 5'-D(*TP*CP*AP*AP*AP*AP*CP*GP*TP*CP*GP*TP*AP *CP*GP*AP*CP*GP*TP*TP*TP*TP*GP*A)-3', DNA ENDONUCLEASE I-CVUI, ... | | Authors: | Molina, R, Redondo, P, LopezMendez, B, Villate, M, Merino, N, Blanco, F.J, Valton, J, Grizot, S, Duchateau, P, Prieto, J, Montoya, G. | | Deposit date: | 2015-07-03 | | Release date: | 2015-09-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of the Homing Endonuclease I-Cvui Provides a New Template for Genome Modification
J.Biol.Chem., 290, 2015
|
|
5A72
 
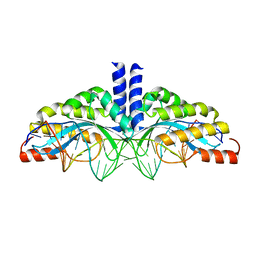 | | Crystal structure of the homing endonuclease I-CvuI in complex with its target (Sro1.3) in the presence of 2 mM Ca | | Descriptor: | 24MER DNA, 5'-D(*DTP*CP*AP*GP*AP*AP*CP*GP*TP*CP*GP*TP*AP *DCP*GP*AP*CP*GP*TP*TP*CP*TP*GP*A)-3', CALCIUM ION, ... | | Authors: | Molina, R, Redondo, P, LopezMendez, B, Villate, M, Merino, N, Blanco, F.J, Valton, J, Grizot, S, Duchateau, P, Prieto, J, Montoya, G. | | Deposit date: | 2015-07-02 | | Release date: | 2015-09-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of the Homing Endonuclease I-Cvui Provides a New Template for Genome Modification
J.Biol.Chem., 290, 2015
|
|
2XAL
 
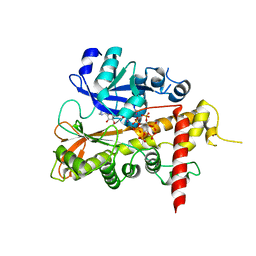 | | Lead derivative of Inositol 1,3,4,5,6-pentakisphosphate 2-kinase from A. thaliana in complex with ADP and IP6. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, INOSITOL-PENTAKISPHOSPHATE 2-KINASE, ... | | Authors: | Gonzalez, B, Banos-Sanz, J.I, Villate, M, Brearley, C.A, Sanz-Aparicio, J. | | Deposit date: | 2010-03-31 | | Release date: | 2010-05-19 | | Last modified: | 2020-10-07 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Inositol 1,3,4,5,6-Pentakisphosphate 2-Kinase is a Distant Ipk Member with a Singular Inositide Binding Site for Axial 2-Oh Recognition.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2XAR
 
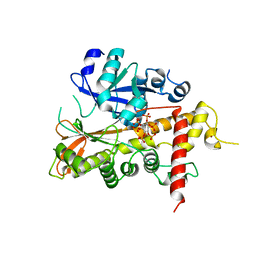 | | Inositol 1,3,4,5,6-pentakisphosphate 2-kinase from A. thaliana in complex with IP6. | | Descriptor: | INOSITOL HEXAKISPHOSPHATE, INOSITOL-PENTAKISPHOSPHATE 2-KINASE, ZINC ION | | Authors: | Gonzalez, B, Banos-Sanz, J.I, Villate, M, Brearley, C.A, Sanz-Aparicio, J. | | Deposit date: | 2010-03-31 | | Release date: | 2010-05-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Inositol 1,3,4,5,6-Pentakisphosphate 2-Kinase is a Distant Ipk Member with a Singular Inositide Binding Site for Axial 2-Oh Recognition.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2XAO
 
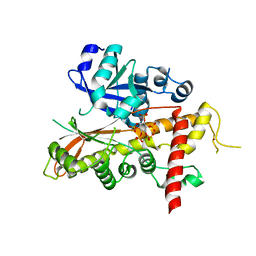 | | Inositol 1,3,4,5,6-pentakisphosphate 2-kinase from A. thaliana in complex with IP5 | | Descriptor: | INOSITOL-PENTAKISPHOSPHATE 2-KINASE, MYO-INOSITOL-(1,3,4,5,6)-PENTAKISPHOSPHATE, ZINC ION | | Authors: | Gonzalez, B, Banos-Sanz, J.I, Villate, M, Brearley, C.A, Sanz-Aparicio, J. | | Deposit date: | 2010-03-31 | | Release date: | 2010-05-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Inositol 1,3,4,5,6-Pentakisphosphate 2-Kinase is a Distant Ipk Member with a Singular Inositide Binding Site for Axial 2-Oh Recognition.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2XAN
 
 | | inositol 1,3,4,5,6-pentakisphosphate 2-kinase from A. thaliana in complex with AMP PNP and IP5 | | Descriptor: | INOSITOL-PENTAKISPHOSPHATE 2-KINASE, MAGNESIUM ION, MYO-INOSITOL-(1,3,4,5,6)-PENTAKISPHOSPHATE, ... | | Authors: | Gonzalez, B, Banos-Sanz, J.I, Villate, M, Brearley, C.A, Sanz-Aparicio, J. | | Deposit date: | 2010-03-31 | | Release date: | 2010-05-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Inositol 1,3,4,5,6-Pentakisphosphate 2-Kinase is a Distant Ipk Member with a Singular Inositide Binding Site for Axial 2-Oh Recognition.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
