3GH0
 
 | | Replacement of Val3 in Human Thymidylate Synthase Affects Its Kinetic Properties and Intracellular Stability | | Descriptor: | SULFATE ION, Thymidylate synthase | | Authors: | Huang, X, Gibson, L.M, Bell, B.J, Lovelace, L.L, Lebioda, L. | | Deposit date: | 2009-03-02 | | Release date: | 2010-03-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Replacement of Val3 in human thymidylate synthase affects its kinetic properties and intracellular stability .
Biochemistry, 49, 2010
|
|
3GH2
 
 | | Replacement of Val3 in Human Thymidylate Synthase Affects Its Kinetic Properties and Intracellular Stability | | Descriptor: | SULFATE ION, Thymidylate synthase | | Authors: | Huang, X, Gibson, L.M, Bell, B.J, Lovelace, L.L, Lebioda, L. | | Deposit date: | 2009-03-02 | | Release date: | 2010-03-02 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Replacement of Val3 in human thymidylate synthase affects its kinetic properties and intracellular stability .
Biochemistry, 49, 2010
|
|
3GG5
 
 | | Replacement of Val3 in Human Thymidylate Synthase Affects Its Kinetic Properties and Intracellular Stability | | Descriptor: | PHOSPHATE ION, SULFATE ION, Thymidylate synthase | | Authors: | Huang, X, Gibson, L.M, Bell, B.J, Lovelace, L.L, Lebioda, L. | | Deposit date: | 2009-02-27 | | Release date: | 2010-03-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Replacement of Val3 in human thymidylate synthase affects its kinetic properties and intracellular stability .
Biochemistry, 49, 2010
|
|
3L9M
 
 | | Crystal structure of PKAB3 (pka triple mutant V123A, L173M, Q181K) with compound 18 | | Descriptor: | (2S)-N~1~-[5-(3-methyl-1H-indazol-5-yl)-1,3,4-thiadiazol-2-yl]-3-(4-methylphenyl)propane-1,2-diamine, cAMP-dependent protein kinase catalytic subunit alpha, cAMP-dependent protein kinase inhibitor alpha | | Authors: | Huang, X. | | Deposit date: | 2010-01-05 | | Release date: | 2011-01-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Azole-based inhibitors of AKT/PKB for the treatment of cancer.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3EJL
 
 | | Replacement of Val3 in Human Thymidylate Synthase Affects Its Kinetic Properties and Intracellular Stability | | Descriptor: | 5-FLUORO-2'-DEOXYURIDINE-5'-MONOPHOSPHATE, PHOSPHATE ION, Thymidylate synthase | | Authors: | Huang, X, Gibson, L.M, Bell, B.J, Lovelace, L.L, Pena, M.M, Berger, F.G, Berger, S.H. | | Deposit date: | 2008-09-18 | | Release date: | 2010-03-02 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Replacement of Val3 in human thymidylate synthase affects its kinetic properties and intracellular stability .
Biochemistry, 49, 2010
|
|
3O89
 
 | | Crystal Structure of Sperm Whale Myoglobin G65T | | Descriptor: | Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Huang, X, Lovelace, L, Lebioda, L. | | Deposit date: | 2010-08-02 | | Release date: | 2011-12-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Investigations of Structural Factors that Influence the Mechanism of Halophenol Dehalogenation using 'Peroxidase-Like' Myoglobin mutants and 'Myoglobin-Like' Amphitrite ornata Dehaloperoxidase Mutants
TO BE PUBLISHED
|
|
3L9N
 
 | | crystal structure of PKAB3 (pka triple mutant V123A, L173M, Q181K) with compound 27 | | Descriptor: | (2S)-N~1~-[5-(1H-indazol-5-yl)-1,3,4-thiadiazol-2-yl]-3-(4-methylphenyl)propane-1,2-diamine, cAMP-dependent protein kinase catalytic subunit alpha, cAMP-dependent protein kinase inhibitor alpha | | Authors: | Huang, X. | | Deposit date: | 2010-01-05 | | Release date: | 2011-01-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Azole-based inhibitors of AKT/PKB for the treatment of cancer.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3L9L
 
 | | Crystal structure of pka with compound 36 | | Descriptor: | 5-[2-({(2S)-2-amino-3-[4-(trifluoromethyl)phenyl]propyl}amino)-1,3-thiazol-5-yl]-1,3-dihydro-2H-indol-2-one, cAMP-dependent protein kinase catalytic subunit alpha, cAMP-dependent protein kinase inhibitor alpha | | Authors: | Huang, X. | | Deposit date: | 2010-01-05 | | Release date: | 2011-01-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Azole-based inhibitors of AKT/PKB for the treatment of cancer.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
4JV7
 
 | | Co-crystal structure of MDM2 with inhibitor (2S,5R,6S)-2-benzyl-5,6-bis(4-bromophenyl)-4-methylmorpholin-3-one | | Descriptor: | (2S,5R,6S)-2-benzyl-5,6-bis(4-bromophenyl)-4-methylmorpholin-3-one, E3 ubiquitin-protein ligase Mdm2, SULFATE ION | | Authors: | Huang, X, Gonzalez-Lopez de Turiso, F, Sun, D, Yosup, R, Bartberger, M.D, Beck, H.P, Cannon, J, Shaffer, P, Oliner, J.D, Olson, S.H, Medina, J.C. | | Deposit date: | 2013-03-25 | | Release date: | 2013-05-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Rational Design and Binding Mode Duality of MDM2-p53 Inhibitors.
J.Med.Chem., 56, 2013
|
|
4JVR
 
 | | Co-crystal structure of MDM2 with inhibitor (2'S,3R,4'S,5'R)-N-(2-aminoethyl)-6-chloro-4'-(3-chloro-2-fluorophenyl)-2'-(2,2-dimethylpropyl)-2-oxo-1,2-dihydrospiro[indole-3,3'-pyrrolidine]-5'-carboxamide | | Descriptor: | (2'S,3R,4'S,5'R)-N-(2-aminoethyl)-6-chloro-4'-(3-chloro-2-fluorophenyl)-2'-(2,2-dimethylpropyl)-2-oxo-1,2-dihydrospiro[indole-3,3'-pyrrolidine]-5'-carboxamide, E3 ubiquitin-protein ligase Mdm2 | | Authors: | Huang, X, Gonzalez-Lopez de Turiso, F, Sun, D, Yosup, R, Bartberger, M.D, Beck, H.P, Cannon, J, Shaffer, P, Oliner, J.D, Olson, S.H, Medina, J.C. | | Deposit date: | 2013-03-26 | | Release date: | 2013-05-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Rational Design and Binding Mode Duality of MDM2-p53 Inhibitors.
J.Med.Chem., 56, 2013
|
|
4JV9
 
 | | Co-crystal structure of MDM2 with inhibitor (2S,5R,6S)-2-benzyl-5,6-bis(4-chlorophenyl)-4-methylmorpholin-3-one | | Descriptor: | (2S,5R,6S)-2-benzyl-5,6-bis(4-chlorophenyl)-4-methylmorpholin-3-one, E3 ubiquitin-protein ligase Mdm2, SULFATE ION | | Authors: | Huang, X, Gonzalez-Lopez de Turiso, F, Sun, D, Yosup, R, Bartberger, M.D, Beck, H.P, Cannon, J, Shaffer, P, Oliner, J.D, Olson, S.H, Medina, J.C. | | Deposit date: | 2013-03-25 | | Release date: | 2013-05-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Rational Design and Binding Mode Duality of MDM2-p53 Inhibitors.
J.Med.Chem., 56, 2013
|
|
4JVE
 
 | | Co-crystal structure of MDM2 with inhibitor (2R,3E)-2-[(2S,3R,6S)-2,3-bis(4-chlorophenyl)-6-(4-fluorobenzyl)-5-oxomorpholin-4-yl]pent-3-enoic acid | | Descriptor: | (2R,3E)-2-[(2S,3R,6S)-2,3-bis(4-chlorophenyl)-6-(4-fluorobenzyl)-5-oxomorpholin-4-yl]pent-3-enoic acid, E3 ubiquitin-protein ligase Mdm2 | | Authors: | Huang, X, Gonzalez-Lopez de Turiso, F, Sun, D, Yosup, R, Bartberger, M.D, Beck, H.P, Cannon, J, Shaffer, P, Oliner, J.D, Olson, S.H, Medina, J.C. | | Deposit date: | 2013-03-25 | | Release date: | 2013-05-01 | | Last modified: | 2013-06-05 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Rational Design and Binding Mode Duality of MDM2-p53 Inhibitors.
J.Med.Chem., 56, 2013
|
|
4RPV
 
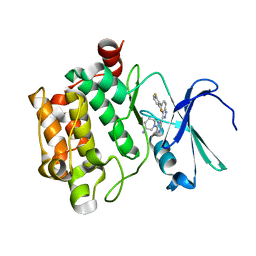 | | co-crystal structure of Pim1 with compound 3 | | Descriptor: | (3S)-1-{6-[5-(2,6-difluorophenyl)-2H-indazol-3-yl]pyrazin-2-yl}piperidin-3-amine, Serine/threonine-protein kinase pim-1 | | Authors: | Huang, X. | | Deposit date: | 2014-10-31 | | Release date: | 2015-02-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | The discovery of novel 3-(pyrazin-2-yl)-1H-indazoles as potent pan-Pim kinase inhibitors.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
3JZK
 
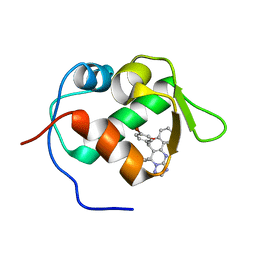 | | crystal structure of MDM2 with chromenotriazolopyrimidine 1 | | Descriptor: | (6R,7S)-6,7-bis(4-bromophenyl)-7,11-dihydro-6H-chromeno[4,3-d][1,2,4]triazolo[1,5-a]pyrimidine, E3 ubiquitin-protein ligase Mdm2 | | Authors: | Huang, X. | | Deposit date: | 2009-09-23 | | Release date: | 2009-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery and optimization of chromenotriazolopyrimidines as potent inhibitors of the mouse double minute 2-tumor protein 53 protein-protein interaction.
J.Med.Chem., 52, 2009
|
|
4DVI
 
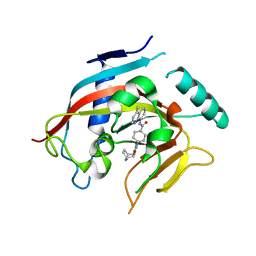 | | Crystal structure of Tankyrase 1 with IWR2 | | Descriptor: | 4-[(3aR,4S,7R,7aS)-1,3-dioxo-1,3,3a,4,7,7a-hexahydro-2H-4,7-methanoisoindol-2-yl]-N-(4-methylquinolin-8-yl)benzamide, Tankyrase-1, ZINC ION | | Authors: | Huang, X. | | Deposit date: | 2012-02-23 | | Release date: | 2012-04-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Novel binding mode of a potent and selective tankyrase inhibitor.
Plos One, 7, 2012
|
|
8IGN
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with RAY1216 | | Descriptor: | (3~{S},3~{a}~{S},6~{a}~{R})-2-[(2~{S})-2-cyclohexyl-2-[2,2,2-tris(fluoranyl)ethanoylamino]ethanoyl]-~{N}-[(2~{S})-4-(cyclopentylamino)-3,4-bis(oxidanylidene)-1-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]butan-2-yl]-3,3~{a},4,5,6,6~{a}-hexahydro-1~{H}-cyclopenta[c]pyrrole-3-carboxamide, 3C-like proteinase nsp5 | | Authors: | Huang, X, Zhou, B, Xu, J, Yang, Z, Zhong, N, Xiong, X. | | Deposit date: | 2023-02-21 | | Release date: | 2023-04-05 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Preclinical evaluation of the SARS-CoV-2 M pro inhibitor RAY1216 shows improved pharmacokinetics compared with nirmatrelvir.
Nat Microbiol, 9, 2024
|
|
8IGO
 
 | | Crystal structure of apo SARS-CoV-2 main protease | | Descriptor: | 3C-like proteinase nsp5 | | Authors: | Huang, X, Zhou, B, Xu, J, Yang, Z, Zhong, N, Xiong, X. | | Deposit date: | 2023-02-21 | | Release date: | 2023-04-05 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Preclinical evaluation of the SARS-CoV-2 M pro inhibitor RAY1216 shows improved pharmacokinetics compared with nirmatrelvir.
Nat Microbiol, 9, 2024
|
|
3QQU
 
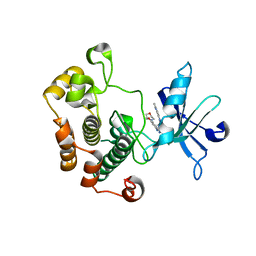 | | Cocrystal structure of unphosphorylated igf with pyrimidine 8 | | Descriptor: | Insulin-like growth factor 1 receptor, N~2~-[3-methoxy-4-(morpholin-4-yl)phenyl]-N~4~-(quinolin-3-yl)pyrimidine-2,4-diamine | | Authors: | Huang, X. | | Deposit date: | 2011-02-16 | | Release date: | 2011-04-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Discovery of 2,4-bis-arylamino-1,3-pyrimidines as insulin-like growth factor-1 receptor (IGF-1R) inhibitors.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
4HNF
 
 | | Crystal structure of ck1d in complex with pf4800567 | | Descriptor: | 3-[(3-chlorophenoxy)methyl]-1-(tetrahydro-2H-pyran-4-yl)-1H-pyrazolo[3,4-d]pyrimidin-4-amine, Casein kinase I isoform delta | | Authors: | Huang, X, Long, A.M, Zhao, H. | | Deposit date: | 2012-10-19 | | Release date: | 2012-11-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structural basis for the potent and selective inhibition of casein kinase 1 epsilon.
J.Med.Chem., 55, 2012
|
|
4HOK
 
 | | crystal structure of apo ck1e | | Descriptor: | Casein kinase I isoform epsilon, SULFATE ION | | Authors: | Huang, X, Long, A.M, Zhao, H. | | Deposit date: | 2012-10-22 | | Release date: | 2012-11-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Structural basis for the potent and selective inhibition of casein kinase 1 epsilon.
J.Med.Chem., 55, 2012
|
|
4HNI
 
 | | crystal structure of ck1e in complex with PF4800567 | | Descriptor: | 3-[(3-chlorophenoxy)methyl]-1-(tetrahydro-2H-pyran-4-yl)-1H-pyrazolo[3,4-d]pyrimidin-4-amine, Casein kinase I isoform epsilon, SULFATE ION | | Authors: | Huang, X, Long, A.M, Zhao, H. | | Deposit date: | 2012-10-19 | | Release date: | 2012-11-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Structural basis for the potent and selective inhibition of casein kinase 1 epsilon.
J.Med.Chem., 55, 2012
|
|
4I9I
 
 | | Crystal structure of tankyrase 1 with compound 4 | | Descriptor: | N-(2-methoxyphenyl)-4-{[3-(4-oxo-3,4-dihydroquinazolin-2-yl)propanoyl]amino}benzamide, Tankyrase-1, ZINC ION | | Authors: | Huang, X. | | Deposit date: | 2012-12-05 | | Release date: | 2013-02-06 | | Last modified: | 2013-05-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery of a class of novel tankyrase inhibitors that bind to both the nicotinamide pocket and the induced pocket.
J.Med.Chem., 56, 2013
|
|
4HBM
 
 | | Ordering of the N Terminus of Human MDM2 by Small Molecule Inhibitors | | Descriptor: | E3 ubiquitin-protein ligase Mdm2, {(3R,5R,6S)-5-(3-chlorophenyl)-6-(4-chlorophenyl)-1-[(2S)-1-hydroxybutan-2-yl]-2-oxopiperidin-3-yl}acetic acid | | Authors: | Huang, X. | | Deposit date: | 2012-09-28 | | Release date: | 2012-10-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Ordering of the N-terminus of human MDM2 by small molecule inhibitors.
J.Am.Chem.Soc., 134, 2012
|
|
3TJC
 
 | | Co-crystal structure of jak2 with thienopyridine 8 | | Descriptor: | 4-amino-N-methyl-2-[4-(morpholin-4-yl)phenyl]thieno[3,2-c]pyridine-7-carboxamide, Tyrosine-protein kinase JAK2 | | Authors: | Huang, X. | | Deposit date: | 2011-08-24 | | Release date: | 2011-11-30 | | Last modified: | 2011-12-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery of potent and highly selective thienopyridine janus kinase 2 inhibitors.
J.Med.Chem., 54, 2011
|
|
3TJD
 
 | | co-crystal structure of Jak2 with thienopyridine 19 | | Descriptor: | 4-amino-2-[4-(tert-butylsulfamoyl)phenyl]-N-methylthieno[3,2-c]pyridine-7-carboxamide, Tyrosine-protein kinase JAK2 | | Authors: | Huang, X. | | Deposit date: | 2011-08-24 | | Release date: | 2011-11-30 | | Last modified: | 2011-12-28 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Discovery of potent and highly selective thienopyridine janus kinase 2 inhibitors.
J.Med.Chem., 54, 2011
|
|
