8UVX
 
 | | CosR DNA bound form I | | Descriptor: | DNA (5'-D(*AP*TP*AP*TP*CP*TP*TP*AP*AP*TP*TP*TP*TP*GP*GP*TP*TP*AP*AP*TP*A)-3'), DNA (5'-D(*TP*AP*TP*TP*AP*AP*CP*CP*AP*AP*AP*AP*TP*TP*AP*AP*GP*AP*TP*AP*T)-3'), DNA-binding response regulator | | Authors: | Zhang, Z. | | Deposit date: | 2023-11-05 | | Release date: | 2024-01-31 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis of DNA recognition of the Campylobacter jejuni CosR regulator.
Mbio, 15, 2024
|
|
5XMC
 
 | |
4OV4
 
 | | Isopropylmalate synthase binding with ketoisovalerate | | Descriptor: | 2-isopropylmalate synthase, 3-METHYL-2-OXOBUTANOIC ACID, ZINC ION | | Authors: | Zhang, Z, Wu, J, Wang, C, Zhang, P. | | Deposit date: | 2014-02-20 | | Release date: | 2014-08-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Subdomain II of alpha-isopropylmalate synthase is essential for activity: inferring a mechanism of feedback inhibition.
J.Biol.Chem., 289, 2014
|
|
2OPW
 
 | | Crystal structure of human phytanoyl-CoA dioxygenase PHYHD1 (apo) | | Descriptor: | PHYHD1 protein | | Authors: | Zhang, Z, Butler, D, McDonough, M.A, Kavanagh, K.L, Bray, J.E, Ng, S.S, von Delft, F, Arrowsmith, C.H, Weigelt, J, Edwards, A, Sundstrom, M, Schofield, C.J, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-01-30 | | Release date: | 2007-03-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of human phytanoyl-CoA dioxygenase PHYHD1 (apo)
To be Published
|
|
2NCA
 
 | |
2QMY
 
 | | Quinone Reductase 2 in complex with adrenochrome | | Descriptor: | (3S)-3-hydroxy-1-methyl-2,3-dihydro-1H-indole-5,6-dione, FLAVIN-ADENINE DINUCLEOTIDE, Ribosyldihydronicotinamide dehydrogenase, ... | | Authors: | Zhang, Z, Fu, Y, Buryanovskky, L. | | Deposit date: | 2007-07-17 | | Release date: | 2008-06-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Quinone Reductase 2 regulates Catecholamine oxidation
To be Published
|
|
7BZH
 
 | |
7CM5
 
 | | Full-length Sarm1 in a self-inhibited state | | Descriptor: | NAD(+) hydrolase SARM1 | | Authors: | Zhang, Z, Jiang, Y. | | Deposit date: | 2020-07-24 | | Release date: | 2020-10-21 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | The NAD + -mediated self-inhibition mechanism of pro-neurodegenerative SARM1.
Nature, 588, 2020
|
|
7CM7
 
 | | NAD+-bound Sarm1 E642A in the self-inhibited state | | Descriptor: | NAD(+) hydrolase SARM1, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Zhang, Z, Jiang, Y. | | Deposit date: | 2020-07-25 | | Release date: | 2020-10-21 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | The NAD + -mediated self-inhibition mechanism of pro-neurodegenerative SARM1.
Nature, 588, 2020
|
|
7CM6
 
 | | NAD+-bound Sarm1 in the self-inhibited state | | Descriptor: | NAD(+) hydrolase SARM1, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Zhang, Z, Jiang, Y. | | Deposit date: | 2020-07-25 | | Release date: | 2020-10-21 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | The NAD + -mediated self-inhibition mechanism of pro-neurodegenerative SARM1.
Nature, 588, 2020
|
|
8T4V
 
 | | Crystal structure of compound 1 bound to K-Ras(G12D) | | Descriptor: | 4-{(1R,5S)-3-[(7P)-7-(8-ethynylnaphthalen-1-yl)-8-fluoro-2-{[(4s,7as)-tetrahydro-1H-pyrrolizin-7a(5H)-yl]methoxy}pyrido[4,3-d]pyrimidin-4-yl]-3,8-diazabicyclo[3.2.1]octan-8-yl}-4-oxobutanoic acid, GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Zhang, Z, Zheng, Q, Guiley, K.Z, Shokat, K.M. | | Deposit date: | 2023-06-10 | | Release date: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Strain-release alkylation of Asp12 enables mutant selective targeting of K-Ras-G12D.
Nat.Chem.Biol., 2024
|
|
8UX2
 
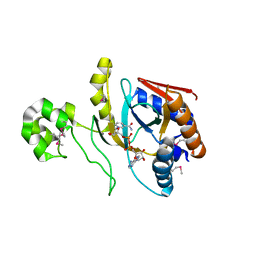 | | Chromobacterium violaceum mono-ADP-ribosyltransferase CteC in complex with NAD+ | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, NAD(+)--protein-threonine ADP-ribosyltransferase, ... | | Authors: | Zhang, Z, Rondon, H, Das, C. | | Deposit date: | 2023-11-08 | | Release date: | 2024-01-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Crystal structure of bacterial ubiquitin ADP-ribosyltransferase CteC reveals a substrate-recruiting insertion.
J.Biol.Chem., 300, 2023
|
|
4QXA
 
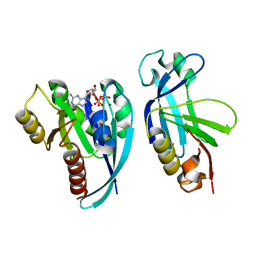 | | Crystal structure of the Rab9A-RUTBC2 RBD complex | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Ras-related protein Rab-9A, ... | | Authors: | Zhang, Z, Wang, S, Ding, J. | | Deposit date: | 2014-07-19 | | Release date: | 2014-09-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the Rab9A-RUTBC2 RBD complex reveals the molecular basis for the binding specificity of Rab9A with RUTBC2.
Structure, 22, 2014
|
|
4N3Y
 
 | |
4N3X
 
 | |
4N3Z
 
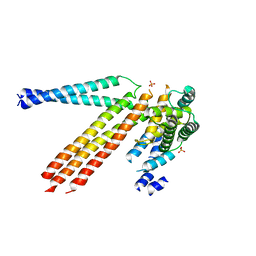 | |
6JCJ
 
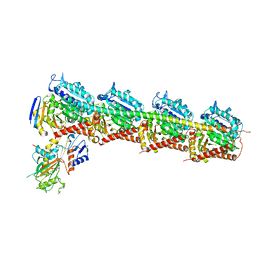 | | Structure of crolibulin in complex with tubulin | | Descriptor: | (4R)-2,7,8-triamino-4-(3-bromo-4,5-dimethoxyphenyl)-4H-1-benzopyran-3-carbonitrile, CALCIUM ION, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Zhang, Z, Yang, J. | | Deposit date: | 2019-01-29 | | Release date: | 2019-03-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular mechanism of crolibulin in complex with tubulin provides a rationale for drug design.
Biochem. Biophys. Res. Commun., 511, 2019
|
|
6IF5
 
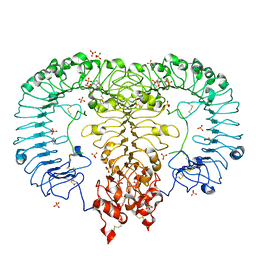 | | Crystal structure of monkey TLR7 in complex with 2',3'-cGMP (Guanosine 2',3'-cyclic phosphate) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GUANOSINE-5'-PHOSPHATE-2',3'-CYCLIC PHOSPHATE, ... | | Authors: | Zhang, Z, Ohto, U, Shimizu, T. | | Deposit date: | 2018-09-18 | | Release date: | 2019-01-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Analyses of Toll-like Receptor 7 Reveal Detailed RNA Sequence Specificity and Recognition Mechanism of Agonistic Ligands.
Cell Rep, 25, 2018
|
|
5YGH
 
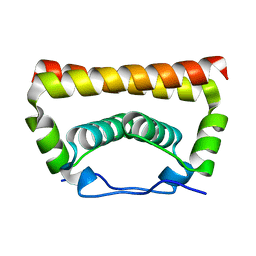 | | Crystal Structure of the Capsid Protein from Zika Virus | | Descriptor: | Capsid protein | | Authors: | Shang, Z, Song, H, Shi, Y, Qi, J, Gao, G.F. | | Deposit date: | 2017-09-23 | | Release date: | 2018-02-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.884 Å) | | Cite: | Crystal Structure of the Capsid Protein from Zika Virus.
J. Mol. Biol., 430, 2018
|
|
7X2Q
 
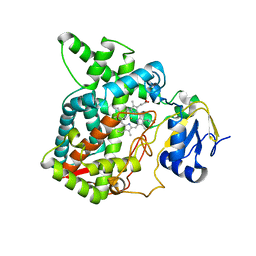 | | Salvia miltiorrhiza CYP76AH3 | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, Sugiol synthase | | Authors: | Chang, Z. | | Deposit date: | 2022-02-26 | | Release date: | 2023-03-01 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.68 Å) | | Cite: | Structure of CYP76AH3 at 3.67 Angstroms resolution
To Be Published
|
|
8SY5
 
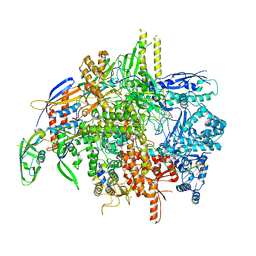 | | E. coli DNA-directed RNA polymerase transcription elongation complex bound the unnatural dS-BTP base pair in the active site | | Descriptor: | 2-oxo-2-hydroadenosine 5'-(tetrahydrogen triphosphate), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Shan, Z, Lyumkis, D, Oh, J, Wang, D. | | Deposit date: | 2023-05-24 | | Release date: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | A unified Watson-Crick geometry drives transcription of six-letter expanded DNA alphabets by E. coli RNA polymerase.
Nat Commun, 14, 2023
|
|
8SY6
 
 | | E. coli DNA-directed RNA polymerase transcription elongation complex bound the unnatural dB-UTP base pair in the active site | | Descriptor: | 2'-DEOXYGUANOSINE-5'-MONOPHOSPHATE, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Shan, Z, Lyumkis, D, Oh, J, Wang, D. | | Deposit date: | 2023-05-24 | | Release date: | 2023-12-27 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | A unified Watson-Crick geometry drives transcription of six-letter expanded DNA alphabets by E. coli RNA polymerase.
Nat Commun, 14, 2023
|
|
8SY7
 
 | | E. coli DNA-directed RNA polymerase transcription elongation complex bound the unnatural dB-STP base pair in the active site | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Shan, Z, Lyumkis, D, Oh, J, Wang, D. | | Deposit date: | 2023-05-24 | | Release date: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (2.65 Å) | | Cite: | A unified Watson-Crick geometry drives transcription of six-letter expanded DNA alphabets by E. coli RNA polymerase.
Nat Commun, 14, 2023
|
|
7XLB
 
 | |
7WGE
 
 | | Human NLRP1 complexed with thioredoxin | | Descriptor: | MAGNESIUM ION, NACHT, LRR and PYD domains-containing protein 1, ... | | Authors: | Zhang, Z, Ohto, U, Shimizu, T. | | Deposit date: | 2021-12-28 | | Release date: | 2023-07-05 | | Last modified: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis for thioredoxin-mediated suppression of NLRP1 inflammasome.
Nature, 622, 2023
|
|
