6XXM
 
 | | Crystal Structure of Human Deoxyhypusine Synthase in complex with putrescine | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,4-DIAMINOBUTANE, Deoxyhypusine synthase | | Authors: | Wator, E, Wilk, P, Grudnik, P. | | Deposit date: | 2020-01-27 | | Release date: | 2020-04-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Half Way to Hypusine-Structural Basis for Substrate Recognition by Human Deoxyhypusine Synthase.
Biomolecules, 10, 2020
|
|
6XXJ
 
 | | Crystal Structure of Human Deoxyhypusine Synthase in complex with spermidine and NAD | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 1,2-ETHANEDIOL, ACETATE ION, ... | | Authors: | Wator, E, Wilk, P, Grudnik, P. | | Deposit date: | 2020-01-27 | | Release date: | 2020-04-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Half Way to Hypusine-Structural Basis for Substrate Recognition by Human Deoxyhypusine Synthase.
Biomolecules, 10, 2020
|
|
6XXH
 
 | | Crystal Structure of Human Deoxyhypusine Synthase in apo form | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, ACETATE ION, BETA-MERCAPTOETHANOL, ... | | Authors: | Wator, E, Wilk, P, Grudnik, P. | | Deposit date: | 2020-01-27 | | Release date: | 2020-04-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Half Way to Hypusine-Structural Basis for Substrate Recognition by Human Deoxyhypusine Synthase.
Biomolecules, 10, 2020
|
|
6XXL
 
 | | Crystal Structure of Human Deoxyhypusine Synthase in complex with spermine | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, ACETATE ION, ... | | Authors: | Wator, E, Wilk, P, Grudnik, P. | | Deposit date: | 2020-01-27 | | Release date: | 2020-04-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Half Way to Hypusine-Structural Basis for Substrate Recognition by Human Deoxyhypusine Synthase.
Biomolecules, 10, 2020
|
|
5NIF
 
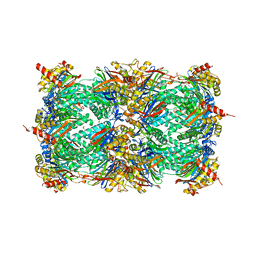 | | Yeast 20S proteasome in complex with Blm-pep activator | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Witkowska, J, Grudnik, P, Golik, P, Dubin, G, Jankowska, E. | | Deposit date: | 2017-03-23 | | Release date: | 2017-08-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of a low molecular weight activator Blm-pep with yeast 20S proteasome - insights into the enzyme activation mechanism.
Sci Rep, 7, 2017
|
|
5OD2
 
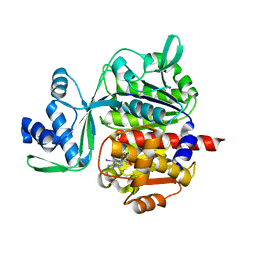 | | Crystal structure of ADP-dependent glucokinase from Methanocaldococcus jannaschii | | Descriptor: | (2R,3R,4S,5R)-2-(4-AMINO-5-IODO-7H-PYRROLO[2,3-D]PYRIMIDIN-7-YL)-5-(HYDROXYMETHYL)TETRAHYDROFURAN-3,4-DIOL, Bifunctional ADP-specific glucokinase/phosphofructokinase, MAGNESIUM ION, ... | | Authors: | Wisniewska, M, Tokarz, P, Grudnik, P. | | Deposit date: | 2017-07-04 | | Release date: | 2018-01-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Crystal structure of ADP-dependent glucokinase from Methanocaldococcus jannaschii in complex with 5-iodotubercidin reveals phosphoryl transfer mechanism.
Protein Sci., 27, 2018
|
|
6YCR
 
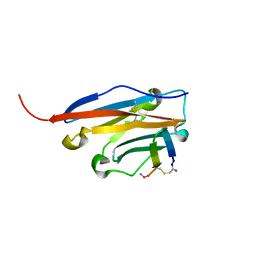 | | Structure of human PD-L1 in complex with inhibitor | | Descriptor: | FFIVIRDRVFR(CCS)G(NH2), Programmed cell death 1 ligand 1 | | Authors: | Magiera-Mularz, K, Grudnik, P, Kuska, K, Holak, T.A, Dubin, G. | | Deposit date: | 2020-03-18 | | Release date: | 2021-02-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Macrocyclic Peptide Inhibitor of PD-1/PD-L1 Immune Checkpoint
Adv. Ther., 2020
|
|
5N2F
 
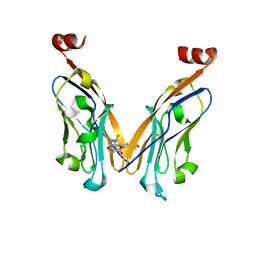 | | Structure of PD-L1/small-molecule inhibitor complex | | Descriptor: | 4-[[4-[[3-(2,3-dihydro-1,4-benzodioxin-6-yl)-2-methyl-phenyl]methoxy]-2,5-bis(fluoranyl)phenyl]methylamino]-3-oxidanylidene-butanoic acid, Programmed cell death 1 ligand 1 | | Authors: | Guzik, K, Zak, K.M, Grudnik, P, Dubin, G, Holak, T.A. | | Deposit date: | 2017-02-07 | | Release date: | 2017-06-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Small-Molecule Inhibitors of the Programmed Cell Death-1/Programmed Death-Ligand 1 (PD-1/PD-L1) Interaction via Transiently Induced Protein States and Dimerization of PD-L1.
J. Med. Chem., 60, 2017
|
|
5N86
 
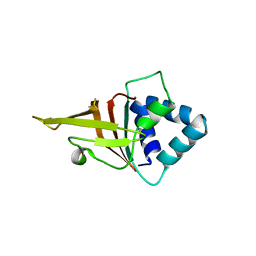 | | Crystal structure of FAS1 domain of hyaluronic acid receptor stabilin-2 | | Descriptor: | Stabilin-2 | | Authors: | Twarda-Clapa, A, Labuzek, B, Grudnik, P, Dubin, G, Holak, T.A. | | Deposit date: | 2017-02-23 | | Release date: | 2018-06-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.484 Å) | | Cite: | Crystal structure of the FAS1 domain of the hyaluronic acid receptor stabilin-2.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
5NIU
 
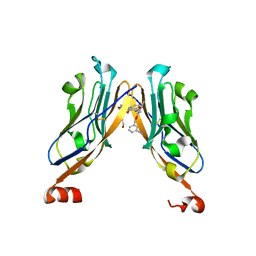 | | Structure of human Programmed cell death 1 ligand 1 (PD-L1) with low molecular mass inhibitor | | Descriptor: | (2~{R})-2-[[2-[(3-cyanophenyl)methoxy]-4-[[3-(2,3-dihydro-1,4-benzodioxin-6-yl)-2-methyl-phenyl]methoxy]-5-methyl-phenyl]methylamino]-3-oxidanyl-propanoic acid, 1,2-ETHANEDIOL, Programmed cell death 1 ligand 1 | | Authors: | Zak, K.M, Grudnik, P, Skalniak, L, Dubin, G, Holak, T.A. | | Deposit date: | 2017-03-27 | | Release date: | 2017-12-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Small-molecule inhibitors of PD-1/PD-L1 immune checkpoint alleviate the PD-L1-induced exhaustion of T-cells.
Oncotarget, 8, 2017
|
|
5O4Y
 
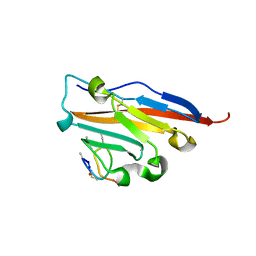 | | Structure of human PD-L1 in complex with inhibitor | | Descriptor: | PHE-MAA-ASN-PRO-HIS-LEU-SER-TRP-SER-TRP-9KK-9KK-ARG-CCS-GLY-NH2, Programmed cell death 1 ligand 1 | | Authors: | Magiera, K, Grudnik, P, Dubin, G, Holak, T.A. | | Deposit date: | 2017-05-31 | | Release date: | 2017-09-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Bioactive Macrocyclic Inhibitors of the PD-1/PD-L1 Immune Checkpoint.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
5N2D
 
 | | Structure of PD-L1/small-molecule inhibitor complex | | Descriptor: | Programmed cell death 1 ligand 1, ~{N}-[2-[[2,6-dimethoxy-4-[(2-methyl-3-phenyl-phenyl)methoxy]phenyl]methylamino]ethyl]ethanamide | | Authors: | Guzik, K, Zak, K.M, Grudnik, P, Dubin, G, Holak, T.A. | | Deposit date: | 2017-02-07 | | Release date: | 2017-06-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Small-Molecule Inhibitors of the Programmed Cell Death-1/Programmed Death-Ligand 1 (PD-1/PD-L1) Interaction via Transiently Induced Protein States and Dimerization of PD-L1.
J. Med. Chem., 60, 2017
|
|
5O45
 
 | | Structure of human PD-L1 in complex with inhibitor | | Descriptor: | PHE-MEA-9KK-SAR-ASP-VAL-MEA-TYR-SAR-TRP-TYR-LEU-CCS-GLY-NH2, Programmed cell death 1 ligand 1 | | Authors: | Magiera, K, Grudnik, P, Dubin, G, Holak, T.A. | | Deposit date: | 2017-05-26 | | Release date: | 2017-09-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (0.99 Å) | | Cite: | Bioactive Macrocyclic Inhibitors of the PD-1/PD-L1 Immune Checkpoint.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
7NLD
 
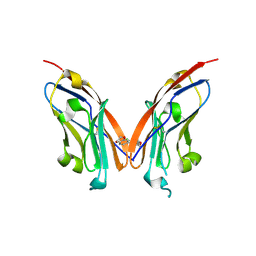 | | Structure of human Programmed cell death 1 ligand 1 (PD-L1) with low molecular mass inhibitor | | Descriptor: | N-(2-((2'-chloro-3'-(2,3-dihydrobenzo[b][1,4]dioxin-6-yl)-3-methoxy-[1,1'-biphenyl]-4-yl)(methyl)amino)ethyl)methanesulfonamide, Programmed cell death 1 ligand 1 | | Authors: | Sala, D, Magiera-Mularz, K, Muszak, D, Surmiak, E, Grudnik, P, Holak, T.A. | | Deposit date: | 2021-02-22 | | Release date: | 2021-08-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Terphenyl-Based Small-Molecule Inhibitors of Programmed Cell Death-1/Programmed Death-Ligand 1 Protein-Protein Interaction.
J.Med.Chem., 64, 2021
|
|
7AD0
 
 | | X-ray structure of Mdm2 with modified p53 peptide | | Descriptor: | E3 ubiquitin-protein ligase Mdm2, Modified p53 peptide | | Authors: | Twarda-Clapa, A, Fortuna, P, Grudnik, P, Dubin, G, Berlicki, L, Holak, T.A. | | Deposit date: | 2020-09-13 | | Release date: | 2020-10-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Systematic ""foldamerization"" of peptide inhibiting p53-MDM2/X interactions by the incorporation of trans- or cis-2-aminocyclopentanecarboxylic acid residues
Eur.J.Med.Chem., 208, 2020
|
|
6R3K
 
 | | Structure of human Programmed cell death 1 ligand 1 (PD-L1) with low molecular mass inhibitor | | Descriptor: | (2~{S},4~{R})-1-[[5-chloranyl-2-[(3-cyanophenyl)methoxy]-4-[[3-(2,3-dihydro-1,4-benzodioxin-6-yl)-2-methyl-phenyl]methoxy]phenyl]methyl]-4-oxidanyl-pyrrolidine-2-carboxylic acid, 1,2-ETHANEDIOL, Programmed cell death 1 ligand 1 | | Authors: | Zak, K.M, Grudnik, P, Skalniak, L, Dubin, G, Holak, T.A. | | Deposit date: | 2019-03-20 | | Release date: | 2019-04-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Terphenyl-Based Small-Molecule Inhibitors of Programmed Cell Death-1/Programmed Death-Ligand 1 Protein-Protein Interaction.
J.Med.Chem., 64, 2021
|
|
5OAI
 
 | | Structure of MDM2 with low molecular weight inhibitor | | Descriptor: | 3-[(1~{R})-2-(~{tert}-butylamino)-1-[methanoyl-[[3,4,5-tris(fluoranyl)phenyl]methyl]amino]-2-oxidanylidene-ethyl]-6-chloranyl-1~{H}-indole-2-carboxylic acid, E3 ubiquitin-protein ligase Mdm2 | | Authors: | Twarda-Clapa, A, Neochoritis, C.G, Grudnik, P, Dubin, G, Domling, A, Holak, T.A. | | Deposit date: | 2017-06-22 | | Release date: | 2019-02-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A fluorinated indole-based MDM2 antagonist selectively inhibits the growth of p53wtosteosarcoma cells.
Febs J., 286, 2019
|
|
