5XEZ
 
 | | Structure of the Full-length glucagon class B G protein-coupled receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-{[(4-cyclohexylphenyl){[3-(methylsulfonyl)phenyl]carbamoyl}amino]methyl}-N-(1H-tetrazol-5-yl)benzamide, ... | | Authors: | Zhang, H, Qiao, A, Yang, D, Yang, L, Dai, A, de Graaf, C, Reedtz-Runge, S, Dharmarajan, V, Zhang, H, Han, G.W, Grant, T, Sierra, R, Weierstall, U, Nelson, G, Liu, W, Wu, Y, Ma, L, Cai, X, Lin, G, Wu, X, Geng, Z, Dong, Y, Song, G, Griffin, P, Lau, J, Cherezov, V, Yang, H, Hanson, M, Stevens, R, Jiang, H, Wang, M, Zhao, Q, Wu, B. | | Deposit date: | 2017-04-06 | | Release date: | 2017-05-24 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of the full-length glucagon class B G-protein-coupled receptor.
Nature, 546, 2017
|
|
2XSE
 
 | | The structural basis for recognition of J-base containing DNA by a novel DNA-binding domain in JBP1 | | Descriptor: | GLYCEROL, NITRATE ION, THYMINE DIOXYGENASE JBP1 | | Authors: | Heidebrecht, T, Christodoulou, E, Chalmers, M.J, Jan, S, ter Riete, B, Grover, R.K, Joosten, R.P, Littler, D, vanLuenen, H, Griffin, P.R, Wentworth, P, Borst, P, Perrakis, A. | | Deposit date: | 2010-09-28 | | Release date: | 2011-03-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Structural Basis for Recognition of Base J Containing DNA by a Novel DNA Binding Domain in Jbp1.
Nucleic Acids Res., 39, 2011
|
|
7KRZ
 
 | | Human mitochondrial LONP1 in complex with Bortezomib | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Endogenous co-purified substrate, ... | | Authors: | Shin, M, Watson, E.R, Song, A.S, Mindrebo, J.T, Novick, S.R, Griffin, P, Wiseman, R.L, Lander, G.C. | | Deposit date: | 2020-11-20 | | Release date: | 2021-02-24 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structures of the human LONP1 protease reveal regulatory steps involved in protease activation.
Nat Commun, 12, 2021
|
|
7KSM
 
 | | Human mitochondrial LONP1 with endogenous substrate | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Lon protease homolog, ... | | Authors: | Shin, M, Watson, E.R, Song, A.S, Mindrebo, J.T, Novick, S.R, Griffin, P, Wiseman, R.L, Lander, G.C. | | Deposit date: | 2020-11-23 | | Release date: | 2020-12-02 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structures of the human LONP1 protease reveal regulatory steps involved in protease activation.
Nat Commun, 12, 2021
|
|
7KSL
 
 | | Substrate-free human mitochondrial LONP1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Lon protease homolog, mitochondrial | | Authors: | Shin, M, Watson, E.R, Song, A.S, Mindrebo, J.T, Novick, S.R, Griffin, P, Wiseman, R.L, Lander, G.C. | | Deposit date: | 2020-11-23 | | Release date: | 2020-12-09 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structures of the human LONP1 protease reveal regulatory steps involved in protease activation.
Nat Commun, 12, 2021
|
|
4ZO1
 
 | | Crystal Structure of the T3-bound TR-beta Ligand-binding Domain in complex with RXR-alpha | | Descriptor: | 3,5,3'TRIIODOTHYRONINE, Nuclear receptor coactivator 2, Retinoic acid receptor RXR-alpha, ... | | Authors: | Bruning, J.B, Kojetin, D.J, Matta-Camacho, E, Hughes, T.S, Srinivasan, S, Nwachukwu, J.C, Cavett, V, Nowak, J, Chalmers, M.J, Marciano, D.P, Kamenecka, T.M, Rance, M, Shulman, A.I, Mangelsdorf, D.J, Griffin, P.R, Nettles, K.W. | | Deposit date: | 2015-05-05 | | Release date: | 2015-09-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.221 Å) | | Cite: | Structural mechanism for signal transduction in RXR nuclear receptor heterodimers.
Nat Commun, 6, 2015
|
|
4ZWJ
 
 | | Crystal structure of rhodopsin bound to arrestin by femtosecond X-ray laser | | Descriptor: | Chimera protein of human Rhodopsin, mouse S-arrestin, and T4 Endolysin | | Authors: | Kang, Y, Zhou, X.E, Gao, X, He, Y, Liu, W, Ishchenko, A, Barty, A, White, T.A, Yefanov, O, Han, G.W, Xu, Q, de Waal, P.W, Ke, J, Tan, M.H.E, Zhang, C, Moeller, A, West, G.M, Pascal, B, Eps, N.V, Caro, L.N, Vishnivetskiy, S.A, Lee, R.J, Suino-Powell, K.M, Gu, X, Pal, K, Ma, J, Zhi, X, Boutet, S, Williams, G.J, Messerschmidt, M, Gati, C, Zatsepin, N.A, Wang, D, James, D, Basu, S, Roy-Chowdhury, S, Conrad, C, Coe, J, Liu, H, Lisova, S, Kupitz, C, Grotjohann, I, Fromme, R, Jiang, Y, Tan, M, Yang, H, Li, J, Wang, M, Zheng, Z, Li, D, Howe, N, Zhao, Y, Standfuss, J, Diederichs, K, Dong, Y, Potter, C.S, Carragher, B, Caffrey, M, Jiang, H, Chapman, H.N, Spence, J.C.H, Fromme, P, Weierstall, U, Ernst, O.P, Katritch, V, Gurevich, V.V, Griffin, P.R, Hubbell, W.L, Stevens, R.C, Cherezov, V, Melcher, K, Xu, H.E, GPCR Network (GPCR) | | Deposit date: | 2015-05-19 | | Release date: | 2015-07-29 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.302 Å) | | Cite: | Crystal structure of rhodopsin bound to arrestin by femtosecond X-ray laser.
Nature, 523, 2015
|
|
7M74
 
 | | ATP-bound AMP-activated protein kinase | | Descriptor: | 5'-AMP-activated protein kinase catalytic subunit alpha-1, 5'-AMP-activated protein kinase subunit beta-2, 5'-AMP-activated protein kinase subunit gamma-1, ... | | Authors: | Yan, Y, Mukherjee, S, Harikumar, K.G, Strutzenberg, T, Zhou, X.E, Powell, S.K, Xu, T, Sheldon, R, Lamp, J, Brunzelle, J.S, Radziwon, K, Ellis, A, Novick, S.J, Vega, I.E, Jones, R, Miller, L.J, Xu, H.E, Griffin, P.R, Kossiakoff, A.A, Melcher, K. | | Deposit date: | 2021-03-26 | | Release date: | 2021-12-15 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.93 Å) | | Cite: | Structure of an AMPK complex in an inactive, ATP-bound state.
Science, 373, 2021
|
|
5TTO
 
 | | X-ray crystal structure of PPARgamma in complex with SR1643 | | Descriptor: | 4-bromo-N-{3,5-dichloro-4-[(quinolin-3-yl)oxy]phenyl}-2,5-difluorobenzene-1-sulfonamide, Peroxisome proliferator-activated receptor gamma | | Authors: | Bruning, J.B, Frkic, R.L, Griffin, P, Kamenecka, T, Abell, A. | | Deposit date: | 2016-11-04 | | Release date: | 2017-05-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.246 Å) | | Cite: | Structure-Activity Relationship of 2,4-Dichloro-N-(3,5-dichloro-4-(quinolin-3-yloxy)phenyl)benzenesulfonamide (INT131) Analogs for PPAR gamma-Targeted Antidiabetics.
J. Med. Chem., 60, 2017
|
|
5XF1
 
 | | Structure of the Full-length glucagon class B G protein-coupled receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, H, Qiao, A, Yang, D, Yang, L, Dai, A, de Graaf, C, Reedtz-Runge, S, Dharmarajan, V, Zhang, H, Han, G.W, Grant, T, Sierra, R, Weierstall, U, Nelson, G, Liu, W, Wu, Y, Ma, L, Cai, X, Lin, G, Wu, X, Geng, Z, Dong, Y, Song, G, Griffin, P, Lau, J, Cherezov, V, Yang, H, Hanson, M, Stevens, R, Jiang, H, Wang, M, Zhao, Q, Wu, B. | | Deposit date: | 2017-04-06 | | Release date: | 2017-05-24 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Structure of the full-length glucagon class B G-protein-coupled receptor.
Nature, 546, 2017
|
|
6M6R
 
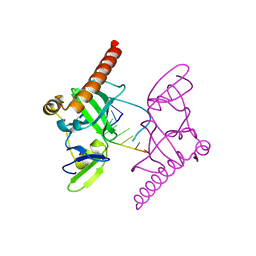 | | Crystal structure of Caenorhabditis elegans Dicer-related helicase 3 (DRH-3) C-terminal domain with 5'-ppp 8-mer ssRNA | | Descriptor: | Dicer Related Helicase, RNA (5'-R(*(GTP)P*GP*CP*CP*GP*CP*CP*C)-3'), ZINC ION | | Authors: | Li, K, Zheng, J, Wirawan, M, Xiong, Z, Fedorova, O, Griffin, P, Plyle, A, Luo, D. | | Deposit date: | 2020-03-16 | | Release date: | 2021-03-17 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Insights into the structure and RNA-binding specificity of Caenorhabditis elegans Dicer-related helicase 3 (DRH-3).
Nucleic Acids Res., 49, 2021
|
|
6M6Q
 
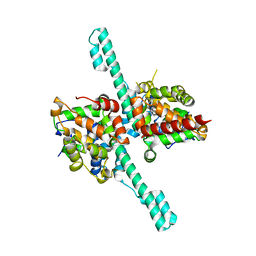 | | Crystal structure of Caenorhabditis elegans Dicer-related helicase 3 (DRH-3) N-terminal domain | | Descriptor: | Dicer Related Helicase | | Authors: | Li, K, Zheng, J, Wirawan, M, Xiong, Z, Fedorova, O, Griffin, P, Plyle, A, Luo, D. | | Deposit date: | 2020-03-16 | | Release date: | 2021-03-17 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Insights into the structure and RNA-binding specificity of Caenorhabditis elegans Dicer-related helicase 3 (DRH-3).
Nucleic Acids Res., 49, 2021
|
|
6M6S
 
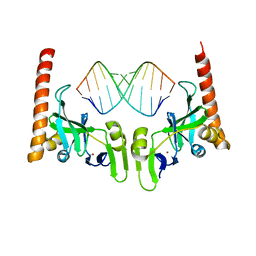 | | Crystal structure of Caenorhabditis elegans Dicer-related helicase 3 (DRH-3) C-terminal domain with 5'-ppp 12-mer dsRNA | | Descriptor: | Dicer Related Helicase, RNA (5'-R(*(GTP)P*GP*CP*GP*CP*GP*CP*GP*CP*GP*CP*C)-3'), ZINC ION | | Authors: | Li, K, Zheng, J, Wirawan, M, Xiong, Z, Fedorova, O, Griffin, P, Plyle, A, Luo, D. | | Deposit date: | 2020-03-16 | | Release date: | 2021-03-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Insights into the structure and RNA-binding specificity of Caenorhabditis elegans Dicer-related helicase 3 (DRH-3).
Nucleic Acids Res., 49, 2021
|
|
4PP6
 
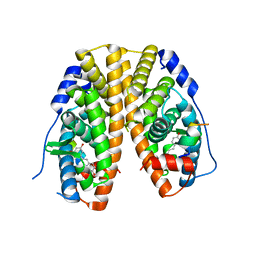 | | Crystal Structure of the Estrogen Receptor alpha Ligand-binding Domain in Complex with Resveratrol | | Descriptor: | Estrogen receptor, Nuclear receptor coactivator 2, RESVERATROL | | Authors: | Nwachukwu, J.C, Srinivasan, S, Bruno, N.E, Parent, A.A, Hughes, T.S, Pollock, J.A, Gjyshi, O, Cavett, V, Nowak, J, Garcia-Ordonez, R.D, Houtman, R, Griffin, P.R, Kojetin, D.J, Katzenellenbogen, J.A, Conkright, M.D, Nettles, K.W. | | Deposit date: | 2014-02-26 | | Release date: | 2014-05-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | Resveratrol modulates the inflammatory response via an estrogen receptor-signal integration network.
Elife, 3, 2014
|
|
4PPS
 
 | | Crystal Structure of the Estrogen Receptor alpha Ligand-binding Domain in Complex with an A-CD ring estrogen derivative | | Descriptor: | (1S,3aR,5R,7aS)-5-(4-hydroxyphenyl)-7a-methyloctahydro-1H-inden-1-ol, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Srinivasan, S, Bruno, N.E, Parent, A.A, Hughes, T.S, Pollock, J.A, Gjyshi, O, Cavett, V, Nowak, J, Garcia-Ordonez, R.D, Houtman, R, Griffin, P.R, Kojetin, D.J, Katzenellenbogen, J.A, Conkright, M.D, Nettles, K.W. | | Deposit date: | 2014-02-27 | | Release date: | 2014-05-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.929 Å) | | Cite: | Resveratrol modulates the inflammatory response via an estrogen receptor-signal integration network.
Elife, 3, 2014
|
|
4PPP
 
 | | Crystal Structure of the Estrogen Receptor alpha Ligand-binding Domain in Complex with Fluoro-Resveratrol | | Descriptor: | 5-[(E)-2-(3-fluoro-4-hydroxyphenyl)ethenyl]benzene-1,3-diol, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Srinivasan, S, Bruno, N.E, Parent, A.A, Hughes, T.S, Pollock, J.A, Gjyshi, O, Cavett, V, Nowak, J, Garcia-Ordonez, R.D, Houtman, R, Griffin, P.R, Kojetin, D.J, Katzenellenbogen, J.A, Conkright, M.D, Nettles, K.W. | | Deposit date: | 2014-02-27 | | Release date: | 2014-05-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.686 Å) | | Cite: | Resveratrol modulates the inflammatory response via an estrogen receptor-signal integration network.
Elife, 3, 2014
|
|
5W0P
 
 | | Crystal structure of rhodopsin bound to visual arrestin determined by X-ray free electron laser | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Endolysin,Rhodopsin,S-arrestin | | Authors: | Zhou, X.E, He, Y, de Waal, P.W, Gao, X, Kang, Y, Van Eps, N, Yin, Y, Pal, K, Goswami, D, White, T.A, Barty, A, Latorraca, N.R, Chapman, H.N, Hubbell, W.L, Dror, R.O, Stevens, R.C, Cherezov, V, Gurevich, V.V, Griffin, P.R, Ernst, O.P, Melcher, K, Xu, H.E. | | Deposit date: | 2017-05-31 | | Release date: | 2017-08-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.013 Å) | | Cite: | Identification of Phosphorylation Codes for Arrestin Recruitment by G Protein-Coupled Receptors.
Cell, 170, 2017
|
|
4R2U
 
 | | Crystal Structure of PPARgamma in complex with SR1664 | | Descriptor: | 4'-[(2,3-dimethyl-5-{[(1S)-1-(4-nitrophenyl)ethyl]carbamoyl}-1H-indol-1-yl)methyl]biphenyl-2-carboxylic acid, Peroxisome proliferator-activated receptor gamma | | Authors: | Marciano, D.P, Kamenecka, T, Griffin, P.R, Bruning, J.B. | | Deposit date: | 2014-08-13 | | Release date: | 2015-07-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Pharmacological repression of PPAR gamma promotes osteogenesis.
Nat Commun, 6, 2015
|
|
4R06
 
 | | Crystal Structure of SR2067 bound to PPARgamma | | Descriptor: | 1-(naphthalen-1-ylsulfonyl)-N-[(1S)-1-phenylpropyl]-1H-indole-5-carboxamide, Peroxisome proliferator-activated receptor gamma, SULFATE ION | | Authors: | Marrewijk, L, Kamenecka, T, Griffin, P.R, Bruning, J.B. | | Deposit date: | 2014-07-30 | | Release date: | 2016-01-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | SR2067 Reveals a Unique Kinetic and Structural Signature for PPAR gamma Partial Agonism.
Acs Chem.Biol., 11, 2016
|
|
5V56
 
 | | 2.9A XFEL structure of the multi-domain human smoothened receptor (with E194M mutation) in complex with TC114 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, FLAVIN MONONUCLEOTIDE, N-methyl-N-[1-[4-(2-methylpyrazol-3-yl)phthalazin-1-yl]piperidin-4-yl]-4-nitro-2-(trifluoromethyl)benzamide, ... | | Authors: | Zhang, X, Zhao, F, Wu, Y, Yang, J, Han, G.W, Zhao, S, Ishchenko, A, Ye, L, Lin, X, Ding, K, Dharmarajan, V, Griffin, P.R, Gati, C, Nelson, G, Hunter, M.S, Hanson, M.A, Cherezov, V, Stevens, R.C, Tan, W, Tao, H, Xu, F. | | Deposit date: | 2017-03-13 | | Release date: | 2017-05-24 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of a multi-domain human smoothened receptor in complex with a super stabilizing ligand.
Nat Commun, 8, 2017
|
|
4R6S
 
 | | Crystal structure of PPARgammma in complex with SR1663 | | Descriptor: | 4'-[(2,3-dimethyl-5-{[(1R)-1-(4-nitrophenyl)ethyl]carbamoyl}-1H-indol-1-yl)methyl]biphenyl-2-carboxylic acid, Peroxisome proliferator-activated receptor gamma | | Authors: | Marciano, D.P, Griffin, P.R, Bruning, J.B. | | Deposit date: | 2014-08-26 | | Release date: | 2015-07-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Pharmacological repression of PPAR gamma promotes osteogenesis.
Nat Commun, 6, 2015
|
|
5V57
 
 | | 3.0A SYN structure of the multi-domain human smoothened receptor in complex with TC114 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, FLAVIN MONONUCLEOTIDE, N-methyl-N-[1-[4-(2-methylpyrazol-3-yl)phthalazin-1-yl]piperidin-4-yl]-4-nitro-2-(trifluoromethyl)benzamide, ... | | Authors: | Zhang, X, Zhao, F, Wu, Y, Yang, J, Han, G.W, Zhao, S, Ishchenko, A, Ye, L, Lin, X, Ding, K, Dharmarajan, V, Griffin, P.R, Gati, C, Nelson, G, Hunter, M.S, Hanson, M.A, Cherezov, V, Stevens, R.C, Tan, W, Tao, H, Xu, F. | | Deposit date: | 2017-03-13 | | Release date: | 2017-05-24 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of a multi-domain human smoothened receptor in complex with a super stabilizing ligand.
Nat Commun, 8, 2017
|
|
6NWS
 
 | | RORgamma Ligand Binding Domain | | Descriptor: | 2-chloro-6-fluoro-N-(1-{[3-(trifluoromethyl)phenyl]sulfonyl}-2,3-dihydro-1H-indol-6-yl)benzamide, Nuclear receptor ROR-gamma | | Authors: | Strutzenberg, T.S, Park, H.J, Griffin, P.R. | | Deposit date: | 2019-02-07 | | Release date: | 2019-07-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | HDX-MS reveals structural determinants for ROR gamma hyperactivation by synthetic agonists.
Elife, 8, 2019
|
|
6NWU
 
 | | RORgamma Ligand Binding Domain | | Descriptor: | 6-[(3,5-dichloropyridin-4-yl)methoxy]-1-{[3-(trifluoromethyl)phenyl]sulfonyl}-2,3-dihydro-1H-indole, Nuclear receptor ROR-gamma | | Authors: | Strutzenberg, T.S, Park, H, Griffin, P.R. | | Deposit date: | 2019-02-07 | | Release date: | 2019-07-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | HDX-MS reveals structural determinants for ROR gamma hyperactivation by synthetic agonists.
Elife, 8, 2019
|
|
6NWT
 
 | | RORgamma Ligand Binding Domain | | Descriptor: | 1,1,1,3,3,3-hexafluoro-2-[2-fluoro-4'-({4-[(pyridin-4-yl)methyl]piperazin-1-yl}methyl)[1,1'-biphenyl]-4-yl]propan-2-ol, Nuclear receptor ROR-gamma | | Authors: | Strutzenberg, T.S, Park, H, Griffin, P.R. | | Deposit date: | 2019-02-07 | | Release date: | 2019-07-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | HDX-MS reveals structural determinants for ROR gamma hyperactivation by synthetic agonists.
Elife, 8, 2019
|
|
