2BE1
 
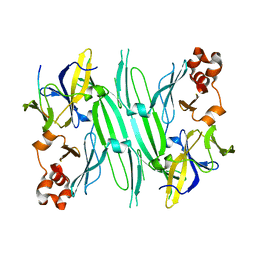 | | Structure of the compact lumenal domain of yeast Ire1 | | Descriptor: | Serine/threonine-protein kinase/endoribonuclease IRE1, peptide | | Authors: | Credle, J.J, Finer-Moore, J.S, Papa, F.R, Stroud, R.M, Walter, P. | | Deposit date: | 2005-10-21 | | Release date: | 2005-12-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.983 Å) | | Cite: | Inaugural Article: On the mechanism of sensing unfolded protein in the endoplasmic reticulum
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
6C9W
 
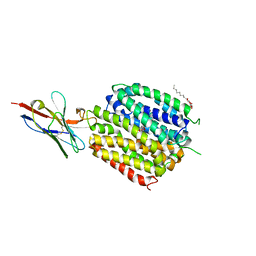 | | Crystal Structure of a ligand bound LacY/Nanobody Complex | | Descriptor: | 4-nitrophenyl alpha-D-galactopyranoside, Lactose permease, Nanobody9047, ... | | Authors: | Kumar, H, Finer-Moore, J.S, Jiang, X, Smirnova, I, Kasho, V, Pardon, E, Steyaert, J, Kaback, H.R, Stroud, R.M. | | Deposit date: | 2018-01-29 | | Release date: | 2018-08-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structure of a ligand-bound LacY-Nanobody Complex.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
1DNA
 
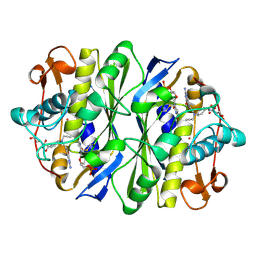 | | D221(169)N MUTANT DOES NOT PROMOTE OPENING OF THE COFACTOR IMIDAZOLIDINE RING | | Descriptor: | 10-PROPARGYL-5,8-DIDEAZAFOLIC ACID, 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, THYMIDYLATE SYNTHASE | | Authors: | Sage, C.R, Michelitsch, M.D, Finer-Moore, J, Stroud, R.M. | | Deposit date: | 1998-06-25 | | Release date: | 1998-11-04 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | D221 in thymidylate synthase controls conformation change, and thereby opening of the imidazolidine.
Biochemistry, 37, 1998
|
|
3FBV
 
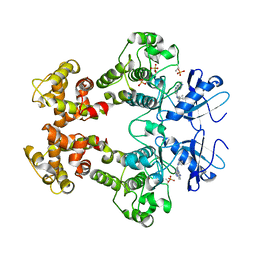 | | Crystal structure of the oligomer formed by the kinase-ribonuclease domain of Ire1 | | Descriptor: | N~2~-1H-benzimidazol-5-yl-N~4~-(3-cyclopropyl-1H-pyrazol-5-yl)pyrimidine-2,4-diamine, Serine/threonine-protein kinase/endoribonuclease IRE1 | | Authors: | Korennykh, A.V, Egea, P.F, Korostelev, A.A, Finer-Moore, J, Zhang, C, Shokat, K.M, Stroud, R.M, Walter, P. | | Deposit date: | 2008-11-19 | | Release date: | 2008-12-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The unfolded protein response signals through high-order assembly of Ire1.
Nature, 457, 2009
|
|
3DH3
 
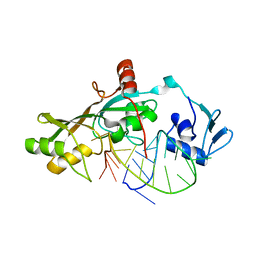 | | Crystal Structure of RluF in complex with a 22 nucleotide RNA substrate | | Descriptor: | Ribosomal large subunit pseudouridine synthase F, stem loop fragment of E. Coli 23S RNA | | Authors: | Alian, A, DeGiovanni, A, Stroud, R.M, Finer-Moore, J.S. | | Deposit date: | 2008-06-16 | | Release date: | 2009-04-28 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of an RluF-RNA complex: a base-pair rearrangement is the key to selectivity of RluF for U2604 of the ribosome.
J.Mol.Biol., 388, 2009
|
|
2OLW
 
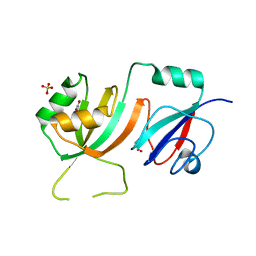 | | Crystal Structure of E. coli pseudouridine synthase RluE | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, ACETIC ACID, Ribosomal large subunit pseudouridine synthase E, ... | | Authors: | Pan, H, Ho, J.D, Stroud, R.M, Finer-Moore, J. | | Deposit date: | 2007-01-19 | | Release date: | 2007-03-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Crystal Structure of E. coli rRNA Pseudouridine Synthase RluE.
J.Mol.Biol., 367, 2007
|
|
4GEV
 
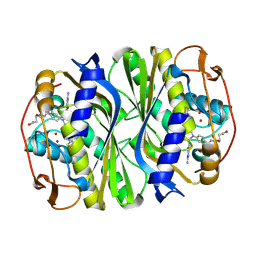 | | E. coli thymidylate synthase Y209W variant in complex with substrate and a cofactor analog | | Descriptor: | 10-PROPARGYL-5,8-DIDEAZAFOLIC ACID, 2'-deoxy-5'-uridylic acid, Thymidylate synthase | | Authors: | Newby, Z, Lee, T.T, Finer-Moore, J, Stroud, R.M. | | Deposit date: | 2012-08-02 | | Release date: | 2012-08-29 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | A remote mutation affects the hydride transfer by disrupting concerted protein motions in thymidylate synthase.
J.Am.Chem.Soc., 134, 2012
|
|
2OML
 
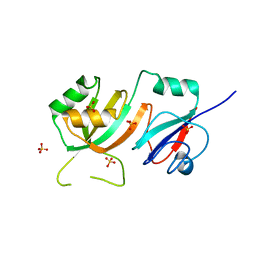 | | crystal structure of E. coli pseudouridine synthase RluE | | Descriptor: | Ribosomal large subunit pseudouridine synthase E, SULFATE ION | | Authors: | Pan, H, Ho, J.D, Stroud, R.M, Finer-Moore, J. | | Deposit date: | 2007-01-22 | | Release date: | 2007-03-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | The Crystal Structure of E. coli rRNA Pseudouridine Synthase RluE.
J.Mol.Biol., 367, 2007
|
|
3R1F
 
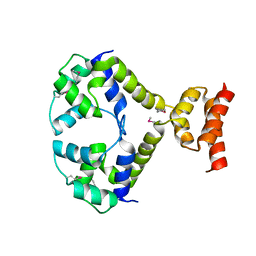 | | Crystal structure of a key regulator of virulence in Mycobacterium tuberculosis | | Descriptor: | ESX-1 secretion-associated regulator EspR | | Authors: | Rosenberg, O.S, Dovey, C, Finer-Moore, J, Stroud, R.M, Cox, J.S. | | Deposit date: | 2011-03-10 | | Release date: | 2011-08-03 | | Last modified: | 2011-08-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | EspR, a key regulator of Mycobacterium tuberculosis virulence, adopts a unique dimeric structure among helix-turn-helix proteins.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3NK5
 
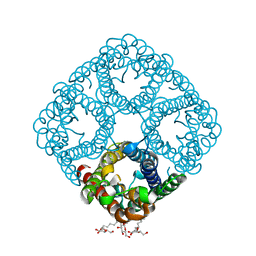 | | Crystal structure of AqpZ mutant F43W | | Descriptor: | Aquaporin Z, octyl beta-D-glucopyranoside | | Authors: | Savage, D.F, O'Connell, J.D, Stroud, R.M, Finer-Moore, J.S. | | Deposit date: | 2010-06-18 | | Release date: | 2010-08-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural context shapes the aquaporin selectivity filter.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2TRM
 
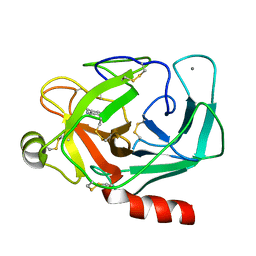 | |
3NKA
 
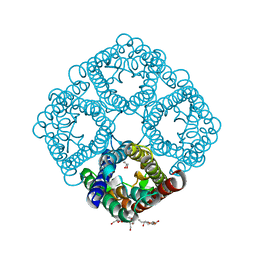 | | Crystal structure of AqpZ H174G,T183F | | Descriptor: | Aquaporin Z, GLYCEROL, octyl beta-D-glucopyranoside | | Authors: | Savage, D.F, O'Connell III, J.D, Finer-Moore, J, Stroud, R.M. | | Deposit date: | 2010-06-18 | | Release date: | 2010-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural context shapes the aquaporin selectivity filter.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
4LXJ
 
 | | Saccharomyces cerevisiae lanosterol 14-alpha demethylase with lanosterol bound | | Descriptor: | LANOSTEROL, Lanosterol 14-alpha demethylase, OXYGEN MOLECULE, ... | | Authors: | Monk, B.C, Tomasiak, T.M, Keniya, M.V, Huschmann, F.U, Tyndall, J.D.A, O'Connell III, J.D, Cannon, R.D, McDonald, J, Rodriguez, A, Finer-Moore, J, Stroud, R.M. | | Deposit date: | 2013-07-29 | | Release date: | 2014-01-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Architecture of a single membrane spanning cytochrome P450 suggests constraints that orient the catalytic domain relative to a bilayer.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4ISK
 
 | | Crystal structure of E.coli thymidylate synthase with dUMP and the BGC 945 inhibitor | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, 2'-deoxy-5'-uridylic acid, MAGNESIUM ION, ... | | Authors: | Tochowicz, A, Finer-Moore, J, Stroud, R.M. | | Deposit date: | 2013-01-16 | | Release date: | 2013-12-25 | | Last modified: | 2014-02-12 | | Method: | X-RAY DIFFRACTION (1.752 Å) | | Cite: | Development and Binding Mode Assessment of N-[4-[2-Propyn-1-yl[(6S)-4,6,7,8-tetrahydro-2-(hydroxymethyl)-4-oxo-3H-cyclopenta[g]quinazolin-6-yl]amino]benzoyl]-l-gamma-glutamyl-d-glutamic Acid (BGC 945), a Novel Thymidylate Synthase Inhibitor That Targets Tumor Cells.
J.Med.Chem., 56, 2013
|
|
3SDJ
 
 | | Structure of RNase-inactive point mutant of oligomeric kinase/RNase Ire1 | | Descriptor: | N~2~-1H-benzimidazol-5-yl-N~4~-(3-cyclopropyl-1H-pyrazol-5-yl)pyrimidine-2,4-diamine, Serine/threonine-protein kinase/endoribonuclease IRE1 | | Authors: | Korennykh, A, Korostelev, A, Egea, P, Finer-Moore, J, Zhang, C, Stroud, R, Shokat, K, Walter, P. | | Deposit date: | 2011-06-09 | | Release date: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.65 Å) | | Cite: | Structural and functional basis for RNA cleavage by Ire1.
Bmc Biol., 9, 2011
|
|
3SDM
 
 | | Structure of oligomeric kinase/RNase Ire1 in complex with an oligonucleotide | | Descriptor: | Serine/threonine-protein kinase/endoribonuclease IRE1 | | Authors: | Korennykh, A, Korostelev, A, Egea, P, Finer-Moore, J, Zhang, C, Stroud, R, Shokat, K, Walter, P. | | Deposit date: | 2011-06-09 | | Release date: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (6.6 Å) | | Cite: | Cofactor-mediated conformational control in the bifunctional kinase/RNase Ire1.
Bmc Biol., 9, 2011
|
|
3NKC
 
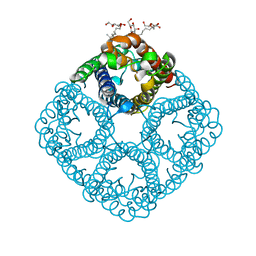 | | Crystal structure of AqpZ F43W,H174G,T183F | | Descriptor: | Aquaporin Z, octyl beta-D-glucopyranoside | | Authors: | Savage, D.F, O'Connell III, J.D, Finer-Moore, J, Stroud, R.M. | | Deposit date: | 2010-06-18 | | Release date: | 2010-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural context shapes the aquaporin selectivity filter.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3NE2
 
 | |
1EXQ
 
 | | CRYSTAL STRUCTURE OF THE HIV-1 INTEGRASE CATALYTIC CORE DOMAIN | | Descriptor: | CADMIUM ION, CHLORIDE ION, POL POLYPROTEIN, ... | | Authors: | Chen, J.C.-H, Krucinski, J, Miercke, L.J.W, Finer-Moore, J.S, Tang, A.H, Leavitt, A.D, Stroud, R.M. | | Deposit date: | 2000-05-03 | | Release date: | 2000-11-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of the HIV-1 integrase catalytic core and C-terminal domains: a model for viral DNA binding.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
4ITS
 
 | | Crystal structure of the catalytic domain of human Pus1 with MES in the active site | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, SULFATE ION, tRNA pseudouridine synthase A, ... | | Authors: | Czudnochowski, N, Finer-Moore, J.S, Stroud, R.M. | | Deposit date: | 2013-01-18 | | Release date: | 2013-06-05 | | Last modified: | 2013-10-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | In Human Pseudouridine Synthase 1 (hPus1), a C-Terminal Helical Insert Blocks tRNA from Binding in the Same Orientation as in the Pus1 Bacterial Homologue TruA, Consistent with Their Different Target Selectivities.
J.Mol.Biol., 425, 2013
|
|
4IQM
 
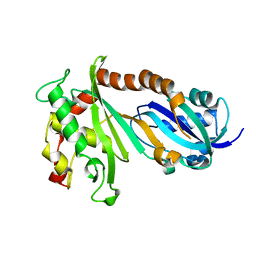 | | Crystal structure of the catalytic domain of human Pus1 | | Descriptor: | tRNA pseudouridine synthase A, mitochondrial | | Authors: | Czudnochowski, N, Finer-Moore, J.S, Stroud, R.M. | | Deposit date: | 2013-01-11 | | Release date: | 2013-06-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | In Human Pseudouridine Synthase 1 (hPus1), a C-Terminal Helical Insert Blocks tRNA from Binding in the Same Orientation as in the Pus1 Bacterial Homologue TruA, Consistent with Their Different Target Selectivities.
J.Mol.Biol., 425, 2013
|
|
4J37
 
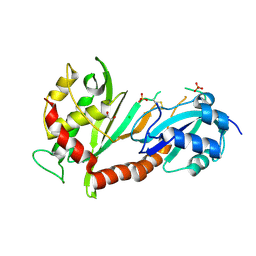 | | Crystal structure of the catalytic domain of human Pus1 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, SULFATE ION, ... | | Authors: | Czudnochowski, N, Finer-Moore, J.S, Stroud, R.M. | | Deposit date: | 2013-02-05 | | Release date: | 2013-06-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | In Human Pseudouridine Synthase 1 (hPus1), a C-Terminal Helical Insert Blocks tRNA from Binding in the Same Orientation as in the Pus1 Bacterial Homologue TruA, Consistent with Their Different Target Selectivities.
J.Mol.Biol., 425, 2013
|
|
1EX4
 
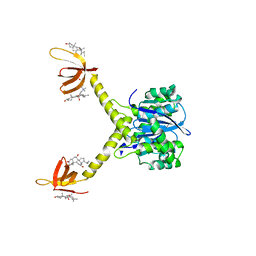 | | HIV-1 INTEGRASE CATALYTIC CORE AND C-TERMINAL DOMAIN | | Descriptor: | 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, INTEGRASE | | Authors: | Chen, J.C.-H, Krucinski, J, Miercke, L.J.W, Finer-Moore, J.S, Tang, A.H, Leavitt, A.D, Stroud, R.M. | | Deposit date: | 2000-04-28 | | Release date: | 2000-06-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the HIV-1 integrase catalytic core and C-terminal domains: a model for viral DNA binding.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
5EQH
 
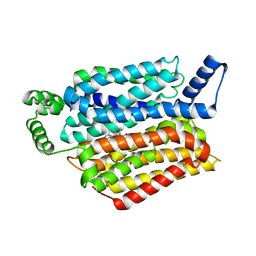 | | Human GLUT1 in complex with inhibitor (2~{S})-3-(2-bromophenyl)-2-[2-(4-methoxyphenyl)ethanoylamino]-~{N}-[(1~{S})-1-phenylethyl]propanamide | | Descriptor: | (2~{S})-3-(2-bromophenyl)-2-[2-(4-methoxyphenyl)ethanoylamino]-~{N}-[(1~{S})-1-phenylethyl]propanamide, Solute carrier family 2, facilitated glucose transporter member 1 | | Authors: | Kapoor, K, Finer-Moore, J, Pedersen, B.P, Caboni, L, Waight, A.B, Hillig, R, Bringmann, P, Heisler, I, Muller, T, Siebeneicher, H, Stroud, R.M. | | Deposit date: | 2015-11-12 | | Release date: | 2016-04-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Mechanism of inhibition of human glucose transporter GLUT1 is conserved between cytochalasin B and phenylalanine amides.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
4E28
 
 | | Structure of human thymidylate synthase in inactive conformation with a novel non-peptidic inhibitor | | Descriptor: | 2-{(2Z,5S)-4-hydroxy-2-[(2E)-(2-hydroxybenzylidene)hydrazinylidene]-2,5-dihydro-1,3-thiazol-5-yl}-N-[3-(trifluoromethyl)phenyl]acetamide, 2-{(5S)-2-[(2E)-2-(2-hydroxybenzylidene)hydrazinyl]-4-oxo-4,5-dihydro-1,3-thiazol-5-yl}-N-[3-(trifluoromethyl)phenyl]acetamide, SULFATE ION, ... | | Authors: | Tochowicz, A, Finer-Moore, J, Stroud, R.M, Costi, M.P. | | Deposit date: | 2012-03-07 | | Release date: | 2012-11-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.302 Å) | | Cite: | Inhibitor of ovarian cancer cells growth by virtual screening: a new thiazole derivative targeting human thymidylate synthase.
J.Med.Chem., 55, 2012
|
|
