6Z69
 
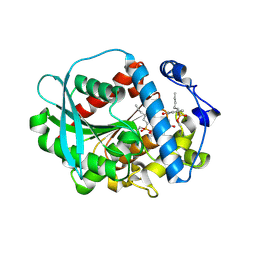 | | A novel metagenomic alpha/beta-fold esterase | | Descriptor: | 7-hydroxy-4-methyl-2H-chromen-2-one, Acetyl esterase/lipase, MAGNESIUM ION, ... | | Authors: | Bollinger, A, Thies, S, Hoeppner, A, Kobus, S, Jaeger, K.-E, Smits, S.H.J. | | Deposit date: | 2020-05-28 | | Release date: | 2020-12-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Crystal structures of a novel family IV esterase in free and substrate-bound form.
Febs J., 288, 2021
|
|
2AF6
 
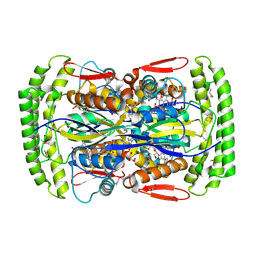 | | Crystal structure of Mycobacterium tuberculosis Flavin dependent thymidylate synthase (Mtb ThyX) in the presence of co-factor FAD and substrate analog 5-Bromo-2'-Deoxyuridine-5'-Monophosphate (BrdUMP) | | Descriptor: | 5-BROMO-2'-DEOXYURIDINE-5'-MONOPHOSPHATE, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Sampathkumar, P, Turley, S, Ulmer, J.E, Rhie, H.G, Sibley, C.H, Hol, W.G. | | Deposit date: | 2005-07-25 | | Release date: | 2005-10-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structure of the Mycobacterium tuberculosis Flavin Dependent Thymidylate Synthase (MtbThyX) at 2.0A Resolution.
J.Mol.Biol., 352, 2005
|
|
1XS4
 
 | | dCTP deaminase from Escherichia coli- E138A mutant enzyme in complex with dCTP | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, Deoxycytidine triphosphate deaminase, MAGNESIUM ION | | Authors: | Johansson, E, Fano, M, Bynck, J.H, Neuhard, J, Larsen, S, Sigurskjold, B.W, Christensen, U, Willemoes, M. | | Deposit date: | 2004-10-18 | | Release date: | 2004-12-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Structures of dCTP deaminase from Escherichia coli with bound substrate and product: reaction mechanism and determinants of mono- and bifunctionality for a family of enzymes
J.Biol.Chem., 280, 2005
|
|
6PXD
 
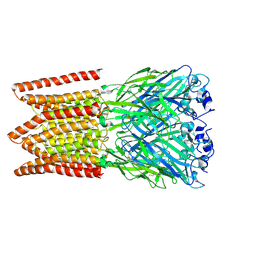 | | CryoEM structure of zebra fish alpha-1 glycine receptor, Apo state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Glycine receptor subunit alphaZ1 | | Authors: | Du, J, Lu, W, Gouaux, E. | | Deposit date: | 2019-07-25 | | Release date: | 2021-02-10 | | Last modified: | 2022-08-24 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Mechanism of gating and partial agonist action in the glycine receptor.
Cell, 184, 2021
|
|
1MU8
 
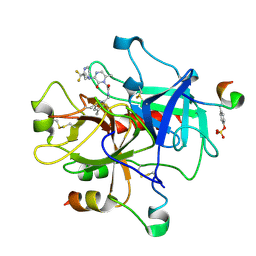 | | thrombin-hirugen_l-378,650 | | Descriptor: | 2-(6-CHLORO-3-{[2,2-DIFLUORO-2-(2-PYRIDINYL)ETHYL]AMINO}-2-OXO-1(2H)-PYRAZINYL)-N-[(2-FLUORO-3-METHYL-6-PYRIDINYL)METHYL]ACETAMIDE, HIRUDIN IIB, THROMBIN | | Authors: | Burgey, C.S, Robinson, K.A, Lyle, T.A, Sanderson, P.E, Lewis, S.D, Lucas, B.J, Krueger, J.A, Singh, R, Miller-Stein, C, White, R.B, Wong, B, Lyle, E.A, Williams, P.D, Coburn, C.A, Dorsey, B.D, Barrow, J.C, Stranieri, M.T, Holahan, M.A, Sitko, G.R, Cook, J.J, McMasters, D.R, McDonough, C.M, Sanders, W.M, Wallace, A.A, Clayton, F.C, Bohn, D, Leonard, Y.M, Detwiler Jr, T.J, Lynch Jr, J.J, Yan, Y, Chen, Z, Kuo, L, Gardell, S.J, Shafer, J.A, Vacca, J.P.J. | | Deposit date: | 2002-09-23 | | Release date: | 2004-04-06 | | Last modified: | 2021-07-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Metabolism-directed optimization of 3-aminopyrazinone acetamide thrombin inhibitors. Development of an orally bioavailable series containing P1 and P3 pyridines.
J.Med.Chem., 46, 2003
|
|
6Z1H
 
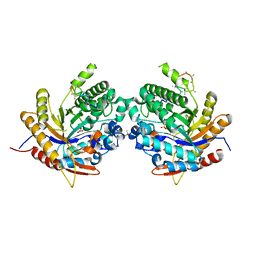 | | Ancestral glycosidase (family 1) | | Descriptor: | ANCESTRAL RECONSTRUCTED GLYCOSIDASE, GLYCEROL, ISOPROPYL ALCOHOL, ... | | Authors: | Gavira, J.A, Risso, V.A, Sanchez-Ruiz, J.M, Gamiz-Arco, G, Gutierrez-Rus, L, Ibarra-Molero, B, Hoshino, Y, Petrovic, D, Romero-Rivera, A, Seelig, B, Kamerlin, S.C.L, Gaucher, E.A. | | Deposit date: | 2020-05-13 | | Release date: | 2020-07-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Heme-binding enables allosteric modulation in an ancient TIM-barrel glycosidase.
Nat Commun, 12, 2021
|
|
5LXK
 
 | | NMR structure of the C-terminal domain of the Bacteriophage T5 decoration protein pb10. | | Descriptor: | Decoration protein | | Authors: | Vernhes, E, Gilquin, B, Cuniasse, P, Boulanger, P, Zinn-Justin, S. | | Deposit date: | 2016-09-22 | | Release date: | 2017-08-02 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | High affinity anchoring of the decoration protein pb10 onto the bacteriophage T5 capsid.
Sci Rep, 7, 2017
|
|
2VRH
 
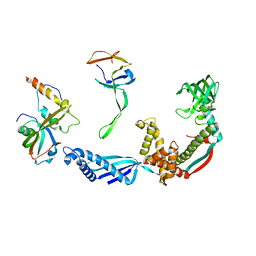 | | Structure of the E. coli trigger factor bound to a translating ribosome | | Descriptor: | 50S RIBOSOMAL PROTEIN L23, 50S RIBOSOMAL PROTEIN L24, 50S RIBOSOMAL PROTEIN L29, ... | | Authors: | Merz, F, Boehringer, D, Schaffitzel, C, Preissler, S, Hoffmann, A, Maier, T, Rutkowska, A, Lozza, J, Ban, N, Bukau, B, Deuerling, E. | | Deposit date: | 2008-04-07 | | Release date: | 2008-06-17 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (19 Å) | | Cite: | Molecular Mechanism and Structure of Trigger Factor Bound to the Translating Ribosome.
Embo J., 27, 2008
|
|
3JC7
 
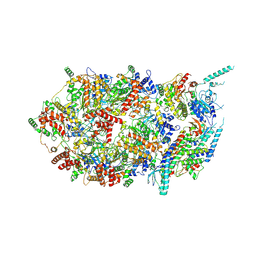 | | Structure of the eukaryotic replicative CMG helicase and pumpjack motion | | Descriptor: | Cell division control protein 45, DNA replication complex GINS protein PSF1, DNA replication complex GINS protein PSF2, ... | | Authors: | Li, H, Bai, L, Yuan, Z, Sun, J, Georgescu, R.E, Liu, J, O'Donnell, M.E. | | Deposit date: | 2015-11-24 | | Release date: | 2016-02-10 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Structure of the eukaryotic replicative CMG helicase suggests a pumpjack motion for translocation.
Nat.Struct.Mol.Biol., 23, 2016
|
|
2VVP
 
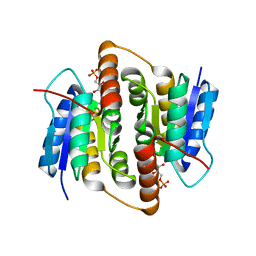 | | Crystal structure of Mycobacterium tuberculosis ribose-5-phosphate isomerase B in complex with its substrates ribose 5-phosphate and ribulose 5-phosphate | | Descriptor: | 5-O-phosphono-D-ribose, RIBOSE-5-PHOSPHATE ISOMERASE B, RIBULOSE-5-PHOSPHATE | | Authors: | Kowalinski, E, Roos, A.K, Mariano, S, Salmon, L, Mowbray, S.L. | | Deposit date: | 2008-06-10 | | Release date: | 2008-07-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | D-Ribose-5-Phosphate Isomerase B from Escherichia Coli is Also a Functional D-Allose-6-Phosphate Isomerase, While the Mycobacterium Tuberculosis Enzyme is not.
J.Mol.Biol., 382, 2008
|
|
4FXI
 
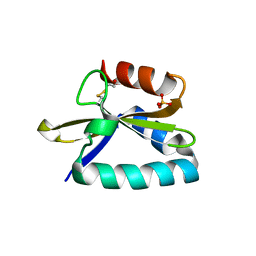 | |
3J9X
 
 | | A Virus that Infects a Hyperthermophile Encapsidates A-Form DNA | | Descriptor: | DNA, coat protein | | Authors: | DiMaio, F, Yu, X, Rensen, E, Krupovic, M, Prangishvili, D, Egelman, E. | | Deposit date: | 2015-03-21 | | Release date: | 2015-06-03 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | A virus that infects a hyperthermophile encapsidates A-form DNA.
Science, 348, 2015
|
|
1U1D
 
 | | Structure of e. coli uridine phosphorylase complexed to 5-(phenylthio)acyclouridine (ptau) | | Descriptor: | 1-((2-HYDROXYETHOXY)METHYL)-5-(PHENYLTHIO)PYRIMIDINE-2,4(1H,3H)-DIONE, PHOSPHATE ION, POTASSIUM ION, ... | | Authors: | Bu, W, Settembre, E.C, Ealick, S.E. | | Deposit date: | 2004-07-15 | | Release date: | 2005-07-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Structural basis for inhibition of Escherichia coli uridine phosphorylase by 5-substituted acyclouridines.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
7KF1
 
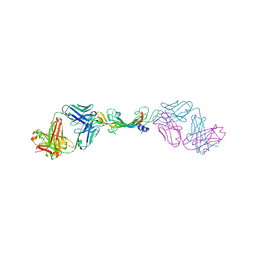 | |
7KGZ
 
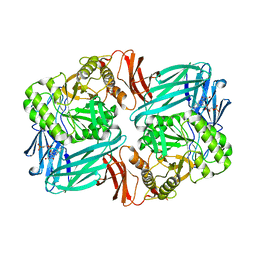 | |
1C7T
 
 | | BETA-N-ACETYLHEXOSAMINIDASE MUTANT E540D COMPLEXED WITH DI-N ACETYL-D-GLUCOSAMINE (CHITOBIASE) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BETA-N-ACETYLHEXOSAMINIDASE, SULFATE ION | | Authors: | Prag, G, Papanikolau, Y, Tavlas, G, Vorgias, C.E, Petratos, K, Oppenheim, A.B. | | Deposit date: | 2000-03-17 | | Release date: | 2000-09-20 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of chitobiase mutants complexed with the substrate Di-N-acetyl-d-glucosamine: the catalytic role of the conserved acidic pair, aspartate 539 and glutamate 540.
J.Mol.Biol., 300, 2000
|
|
6Z8V
 
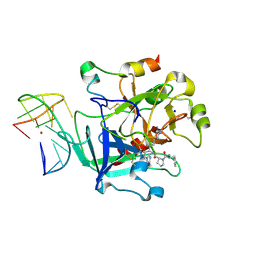 | | X-ray structure of the complex between human alpha thrombin and a thrombin binding aptamer variant (TBA-3L), which contains 1-beta-D-lactopyranosyl residue in the side chain of Thy3 at N3. | | Descriptor: | D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, POTASSIUM ION, Prothrombin, ... | | Authors: | Troisi, R, Timofeev, E.N, Sica, F. | | Deposit date: | 2020-06-02 | | Release date: | 2021-01-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Expanding the recognition interface of the thrombin-binding aptamer HD1 through modification of residues T3 and T12.
Mol Ther Nucleic Acids, 23, 2021
|
|
8BZM
 
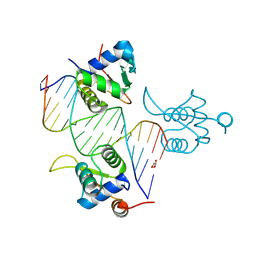 | | FOXK1-ELF1-heterodimer bound to DNA | | Descriptor: | DNA, ETS-related transcription factor Elf-1, Forkhead box protein K1, ... | | Authors: | Morgunova, E, Popov, A, Yin, Y, Taipale, J. | | Deposit date: | 2022-12-15 | | Release date: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | FOXK1-ELF1_heterodimer bound to DNA
To Be Published
|
|
7ZV9
 
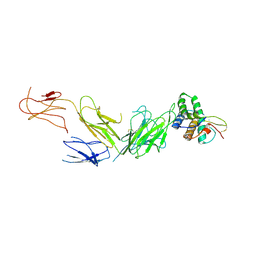 | |
4JI1
 
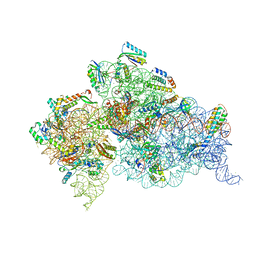 | | Crystal Structure of 30S ribosomal subunit from Thermus thermophilus | | Descriptor: | 16S rRNA, MAGNESIUM ION, RIBOSOMAL PROTEIN S10, ... | | Authors: | Demirci, H, Wang, L, Murphy IV, F, Murphy, E, Carr, J, Blanchard, S, Jogl, G, Dahlberg, A.E, Gregory, S.T. | | Deposit date: | 2013-03-05 | | Release date: | 2013-11-06 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (3.144 Å) | | Cite: | The central role of protein S12 in organizing the structure of the decoding site of the ribosome.
Rna, 19, 2013
|
|
6Z32
 
 | | Human cation-independent mannose 6-phosphate/IGF2 receptor domains 7-11 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Cation-independent mannose-6-phosphate receptor, SULFATE ION, ... | | Authors: | Bochel, A.J, Williams, C, McCoy, A.J, Hoppe, H, Winter, A.J, Nicholls, R.D, Harlos, K, Jones, Y.E, Berger, I, Hassan, B, Crump, M.P. | | Deposit date: | 2020-05-19 | | Release date: | 2020-08-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.47 Å) | | Cite: | Structure of the Human Cation-Independent Mannose 6-Phosphate/IGF2 Receptor Domains 7-11 Uncovers the Mannose 6-Phosphate Binding Site of Domain 9.
Structure, 28, 2020
|
|
6R65
 
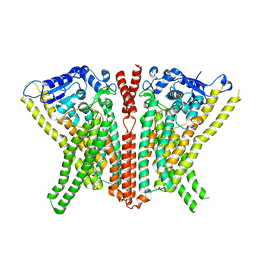 | | Crystal Structure of human TMEM16K / Anoctamin 10 (Form 2) | | Descriptor: | Anoctamin-10, CALCIUM ION | | Authors: | Bushell, S.R, Pike, A.C.W, Chu, A, Tessitore, A, Rotty, B, Mukhopadhyay, S, Kupinska, K, Shrestha, L, Borkowska, O, Chalk, R, Burgess-Brown, N.A, Love, J, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-03-26 | | Release date: | 2019-05-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | The structural basis of lipid scrambling and inactivation in the endoplasmic reticulum scramblase TMEM16K.
Nat Commun, 10, 2019
|
|
6CD7
 
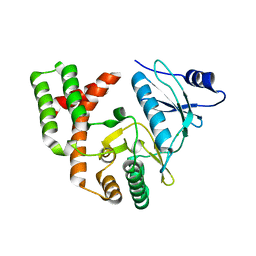 | | Crystal structure of APH(2")-IVa in complex with plazomicin | | Descriptor: | (2S)-4-amino-N-[(1R,2S,3S,4R,5S)-5-amino-4-{[(2S,3R)-3-amino-6-{[(2-hydroxyethyl)amino]methyl}-3,4-dihydro-2H-pyran-2-y l]oxy}-2-{[3-deoxy-4-C-methyl-3-(methylamino)-beta-L-arabinopyranosyl]oxy}-3-hydroxycyclohexyl]-2-hydroxybutanamide, APH(2'')-Id, CHLORIDE ION | | Authors: | Stogios, P.J, Evdokimova, E, Dong, A, Di Leo, R, Savchenko, A, Satchell, K.J, Joachimiak, J, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-02-08 | | Release date: | 2018-02-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Plazomicin Retains Antibiotic Activity against Most Aminoglycoside Modifying Enzymes.
ACS Infect Dis, 4, 2018
|
|
6YCE
 
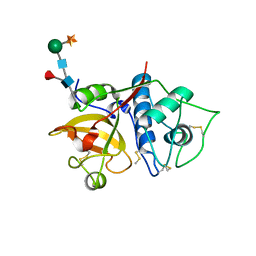 | | Structure the bromelain protease from Ananas comosus with a thiomethylated active cysteine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, FBSB, ISOPROPYL ALCOHOL, ... | | Authors: | Azarkan, M, Charlier, P, Herman, R, Delbrassine, F, Sauvage, E, M Rabet, N, Calvo Esposito, R, Kerff, F. | | Deposit date: | 2020-03-18 | | Release date: | 2020-11-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of the free and inhibitors-bound forms of bromelain and ananain from Ananas comosus stem and in vitro study of their cytotoxicity.
Sci Rep, 10, 2020
|
|
5SVM
 
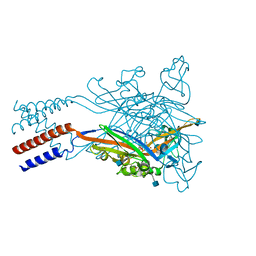 | | Crystal structure of the ATP-gated human P2X3 ion channel bound to agonist 2-methylthio-ATP in the desensitized state | | Descriptor: | 1,2-ETHANEDIOL, 2-(methylsulfanyl)adenosine 5'-(tetrahydrogen triphosphate), 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Mansoor, S.E, Lu, W, Oosterheert, W, Shekhar, M, Tajkhorshid, E, Gouaux, E. | | Deposit date: | 2016-08-06 | | Release date: | 2016-09-28 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.093 Å) | | Cite: | X-ray structures define human P2X3 receptor gating cycle and antagonist action.
Nature, 538, 2016
|
|
