7M2O
 
 | | Crystal structure of Human Glycolate Oxidase with Inhibitor Compound 15 | | Descriptor: | 1,2-ETHANEDIOL, 5-[(5'-{1-(4-carboxy-1,3-thiazol-2-yl)-5-(cyclopropylmethyl)-4-[(3-fluoro-4-sulfamoylphenyl)methyl]-1H-pyrazol-3-yl}-2'-fluoro[1,1'-biphenyl]-4-yl)oxy]-1H-1,2,3-triazole-4-carboxylic acid, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Gumpena, R, Ding, J, Powell, D.A, Lowther, W.T. | | Deposit date: | 2021-03-17 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Dual Glycolate Oxidase/Lactate Dehydrogenase A Inhibitors for Primary Hyperoxaluria
ACS Medicinal Chemistry Letters, 12, 2021
|
|
6V97
 
 | | Kindlin-3 double deletion mutant short form | | Descriptor: | Fermitin family homolog 3 | | Authors: | Xu, Z, Zhang, T.L, Xu, Z, Sun, J.J, Ding, J.P, Ma, Y.Q. | | Deposit date: | 2019-12-13 | | Release date: | 2020-07-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.381 Å) | | Cite: | Structure basis of the FERM domain of kindlin-3 in supporting integrin alpha IIb beta 3 activation in platelets.
Blood Adv, 4, 2020
|
|
6V9G
 
 | | Kindlin-3 double deletion mutant long form | | Descriptor: | Fermitin family homolog 3 | | Authors: | Xu, Z, Zhang, T.L, Xu, Z, Sun, J.J, Ding, J.P, Ma, Y.Q. | | Deposit date: | 2019-12-13 | | Release date: | 2020-07-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure basis of the FERM domain of kindlin-3 in supporting integrin alpha IIb beta 3 activation in platelets.
Blood Adv, 4, 2020
|
|
1UWB
 
 | | TYR 181 CYS HIV-1 RT/8-CL TIBO | | Descriptor: | 5-CHLORO-8-METHYL-7-(3-METHYL-BUT-2-ENYL)-6,7,8,9-TETRAHYDRO-2H-2,7,9A-TRIAZA-BENZO[CD]AZULENE-1-THIONE, REVERSE TRANSCRIPTASE | | Authors: | Das, K, Ding, J, Hsiou, Y, Arnold, E. | | Deposit date: | 1996-11-21 | | Release date: | 1997-05-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structures of 8-Cl and 9-Cl TIBO complexed with wild-type HIV-1 RT and 8-Cl TIBO complexed with the Tyr181Cys HIV-1 RT drug-resistant mutant.
J.Mol.Biol., 264, 1996
|
|
8HQN
 
 | | Activation mechanism of GPR132 by 9(S)-HODE | | Descriptor: | (9S,10E,12Z)-9-hydroxyoctadeca-10,12-dienoic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Wang, J.L, Ding, J.H, Sun, J.P, Yu, X. | | Deposit date: | 2022-12-13 | | Release date: | 2023-10-11 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Functional screening and rational design of compounds targeting GPR132 to treat diabetes.
Nat Metab, 5, 2023
|
|
8HQE
 
 | | Cryo-EM structure of the apo-GPR132-Gi | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Wang, J.L, Ding, J.H, Sun, J.P, Yu, X. | | Deposit date: | 2022-12-13 | | Release date: | 2023-10-11 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Functional screening and rational design of compounds targeting GPR132 to treat diabetes.
Nat Metab, 5, 2023
|
|
8HQM
 
 | | Activation mechanism of GPR132 by NPGLY | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Wang, J.L, Ding, J.H, Sun, J.P, Yu, X. | | Deposit date: | 2022-12-13 | | Release date: | 2023-10-11 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Functional screening and rational design of compounds targeting GPR132 to treat diabetes.
Nat Metab, 5, 2023
|
|
1HNV
 
 | | STRUCTURE OF HIV-1 RT(SLASH)TIBO R 86183 COMPLEX REVEALS SIMILARITY IN THE BINDING OF DIVERSE NONNUCLEOSIDE INHIBITORS | | Descriptor: | 5-CHLORO-8-METHYL-7-(3-METHYL-BUT-2-ENYL)-6,7,8,9-TETRAHYDRO-2H-2,7,9A-TRIAZA-BENZO[CD]AZULENE-1-THIONE, HIV-1 REVERSE TRANSCRIPTASE (SUBUNIT P51), HIV-1 REVERSE TRANSCRIPTASE (SUBUNIT P66) | | Authors: | Das, K, Ding, J, Arnold, E. | | Deposit date: | 1995-03-30 | | Release date: | 1995-07-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of HIV-1 RT/TIBO R 86183 complex reveals similarity in the binding of diverse nonnucleoside inhibitors.
Nat.Struct.Biol., 2, 1995
|
|
1NLN
 
 | | CRYSTAL STRUCTURE OF HUMAN ADENOVIRUS 2 PROTEINASE WITH ITS 11 AMINO ACID COFACTOR AT 1.6 ANGSTROM RESOLUTION | | Descriptor: | ACETIC ACID, Adenain, PVIC | | Authors: | McGrath, W.J, Ding, J, Sweet, R.M, Mangel, W.F. | | Deposit date: | 2003-01-07 | | Release date: | 2003-08-26 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystallographic structure at 1.6-A resolution of the human adenovirus proteinase in a covalent complex with its 11-amino-acid peptide cofactor: insights on a new fold
Biochim.Biophys.Acta, 1648, 2003
|
|
1BQM
 
 | | HIV-1 RT/HBY 097 | | Descriptor: | (S)-4-ISOPROPOXYCARBONYL-6-METHOXY-3-METHYLTHIOMETHYL-3,4-DIHYDROQUINOXALIN-2(1H)-THIONE, REVERSE TRANSCRIPTASE | | Authors: | Hsiou, Y, Das, K, Ding, J, Arnold, E. | | Deposit date: | 1998-08-17 | | Release date: | 1999-01-06 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structures of Tyr188Leu mutant and wild-type HIV-1 reverse transcriptase complexed with the non-nucleoside inhibitor HBY 097: inhibitor flexibility is a useful design feature for reducing drug resistance.
J.Mol.Biol., 284, 1998
|
|
1BQN
 
 | | TYR 188 LEU HIV-1 RT/HBY 097 | | Descriptor: | (S)-4-ISOPROPOXYCARBONYL-6-METHOXY-3-METHYLTHIOMETHYL-3,4-DIHYDROQUINOXALIN-2(1H)-THIONE, REVERSE TRANSCRIPTASE | | Authors: | Hsiou, Y, Das, K, Ding, J, Arnold, E. | | Deposit date: | 1998-08-17 | | Release date: | 1999-01-06 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structures of Tyr188Leu mutant and wild-type HIV-1 reverse transcriptase complexed with the non-nucleoside inhibitor HBY 097: inhibitor flexibility is a useful design feature for reducing drug resistance.
J.Mol.Biol., 284, 1998
|
|
8W78
 
 | | Structure of Drosophila melanogaster L-2-hydroxyglutarate dehydrogenase in complex with FAD and 2-oxoglutarate | | Descriptor: | 2-OXOGLUTARIC ACID, DODECYL-BETA-D-MALTOSIDE, FI05204p, ... | | Authors: | Yang, J, Chen, X, Jin, S, Ding, J. | | Deposit date: | 2023-08-30 | | Release date: | 2023-11-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Structure and biochemical characterization of l-2-hydroxyglutarate dehydrogenase and its role in the pathogenesis of l-2-hydroxyglutaric aciduria.
J.Biol.Chem., 300, 2023
|
|
8W75
 
 | | Structure of Drosophila melanogaster L-2-hydroxyglutarate dehydrogenase | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, FI05204p, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Yang, J, Chen, X, Jin, S, Ding, J. | | Deposit date: | 2023-08-30 | | Release date: | 2023-11-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structure and biochemical characterization of l-2-hydroxyglutarate dehydrogenase and its role in the pathogenesis of l-2-hydroxyglutaric aciduria.
J.Biol.Chem., 300, 2023
|
|
8W7F
 
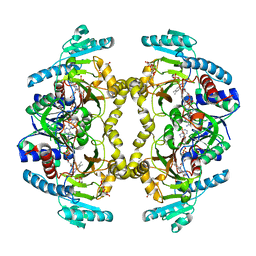 | | Structure of Drosophila melanogaster L-2-hydroxyglutarate dehydrogenase bound with FAD and a sulfate ion | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, FI05204p, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Yang, J, Chen, X, Jin, S, Ding, J. | | Deposit date: | 2023-08-30 | | Release date: | 2023-11-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.299 Å) | | Cite: | Structure and biochemical characterization of l-2-hydroxyglutarate dehydrogenase and its role in the pathogenesis of l-2-hydroxyglutaric aciduria.
J.Biol.Chem., 300, 2023
|
|
1L3K
 
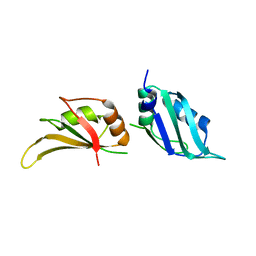 | | UP1, THE TWO RNA-RECOGNITION MOTIF DOMAIN OF HNRNP A1 | | Descriptor: | HETEROGENEOUS NUCLEAR RIBONUCLEOPROTEIN A1 | | Authors: | Vitali, J, Ding, J, Jiang, J, Zhang, Y, Krainer, A.R, Xu, R.-M. | | Deposit date: | 2002-02-27 | | Release date: | 2002-04-17 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Correlated alternative side chain conformations in the RNA-recognition motif of heterogeneous nuclear ribonucleoprotein A1.
Nucleic Acids Res., 30, 2002
|
|
8GS5
 
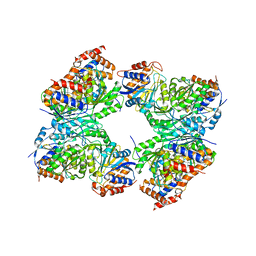 | | Crystal structure of a constitutively active mutant of human IDH3 holoenzyme in apo form | | Descriptor: | Isocitrate dehydrogenase [NAD] subunit alpha, mitochondrial, Isocitrate dehydrogenase [NAD] subunit gamma, ... | | Authors: | Sun, P, Chen, X, Ding, J. | | Deposit date: | 2022-09-04 | | Release date: | 2022-11-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (4.486 Å) | | Cite: | Structures of a constitutively active mutant of human IDH3 reveal new insights into the mechanisms of allosteric activation and the catalytic reaction.
J.Biol.Chem., 298, 2022
|
|
8GRB
 
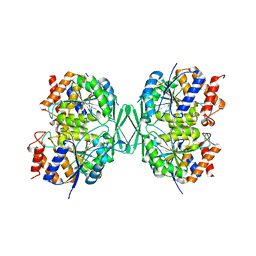 | |
8GTN
 
 | |
5H4B
 
 | | Crystal structure of Cbln4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Cerebellin-4 | | Authors: | Zhong, C, Shen, J, Zhang, H, Ding, J. | | Deposit date: | 2016-10-31 | | Release date: | 2017-09-13 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Cbln1 and Cbln4 Are Structurally Similar but Differ in GluD2 Binding Interactions.
Cell Rep, 20, 2017
|
|
1O5T
 
 | | Crystal structure of the aminoacylation catalytic fragment of human tryptophanyl-tRNA synthetase | | Descriptor: | Tryptophanyl-tRNA synthetase | | Authors: | Yu, Y, Liu, Y, Shen, N, Xu, X, Jia, J, Jin, Y, Arnold, E, Ding, J. | | Deposit date: | 2003-10-05 | | Release date: | 2004-07-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of Human Tryptophanyl-tRNA Synthetase Catalytic Fragment
J.BIOL.CHEM., 279, 2004
|
|
8HVI
 
 | | Activation mechanism of GPR132 by compound NOX-6-7 | | Descriptor: | 3-methyl-5-[(4-oxidanylidene-4-phenyl-butanoyl)amino]-1-benzofuran-2-carboxylic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Wang, J.L, Ding, J.H, Sun, J.P, Yu, X. | | Deposit date: | 2022-12-26 | | Release date: | 2023-10-11 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Functional screening and rational design of compounds targeting GPR132 to treat diabetes.
Nat Metab, 5, 2023
|
|
8GRU
 
 | | Crystal structure of a constitutively active mutant of the alpha beta heterodimer of human IDH3 in complex with ICT, NAD and Ca | | Descriptor: | CALCIUM ION, Human IDH3 alpha subunit, ISOCITRIC ACID, ... | | Authors: | Sun, P, Chen, X, Ding, J. | | Deposit date: | 2022-09-02 | | Release date: | 2022-11-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.847 Å) | | Cite: | Structures of a constitutively active mutant of human IDH3 reveal new insights into the mechanisms of allosteric activation and the catalytic reaction.
J.Biol.Chem., 298, 2022
|
|
8GTJ
 
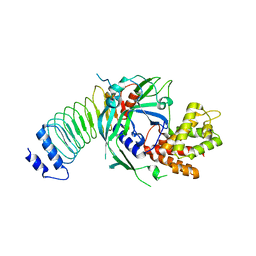 | |
1NFG
 
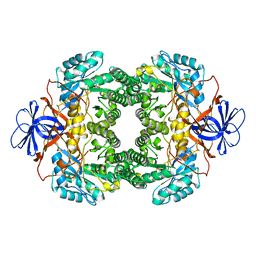 | | Structure of D-hydantoinase | | Descriptor: | D-hydantoinase, ZINC ION | | Authors: | Xu, Z, Yang, Y, Jiang, W, Arnold, E, Ding, J. | | Deposit date: | 2002-12-14 | | Release date: | 2003-07-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of D-Hydantoinase from Burkholderia pickettii at a Resolution of 2.7 Angstroms: Insights into the Molecular Basis of Enzyme Thermostability.
J.Bacteriol., 185, 2003
|
|
5J5C
 
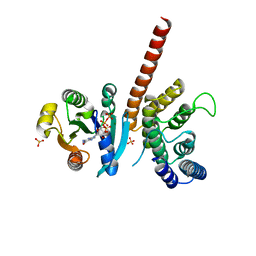 | | Crystal structure of ARL1-GTP and DCB domain of BIG1 complex | | Descriptor: | ADP-ribosylation factor-like protein 1, Brefeldin A-inhibited guanine nucleotide-exchange protein 1, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Wang, R, Wang, Z, Zhang, T, Ding, J. | | Deposit date: | 2016-04-02 | | Release date: | 2016-09-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural basis for targeting BIG1 to Golgi apparatus through interaction of its DCB domain with Arl1
J Mol Cell Biol, 2016
|
|
