2E36
 
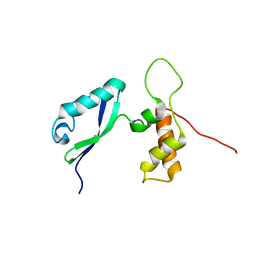 | | L11 with SANS refinement | | Descriptor: | 50S ribosomal protein L11 | | Authors: | Lee, D, Walsh, J.D, Yu, P, Markus, M.A, Choli-Papadopoulous, T, Schwieters, C.D, Krueger, S, Draper, D.E, Wang, Y.X. | | Deposit date: | 2006-11-20 | | Release date: | 2007-06-19 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The structure of free L11 and functional dynamics of L11 in free, L11-rRNA(58 nt) binary and L11-rRNA(58 nt)-thiostrepton ternary complexes
J.Mol.Biol., 367, 2007
|
|
2E4Z
 
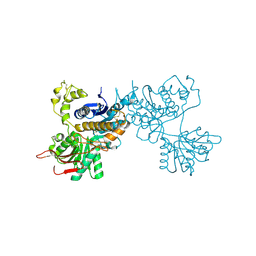 | | Crystal structure of the ligand-binding region of the group III metabotropic glutamate receptor | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Metabotropic glutamate receptor 7 | | Authors: | Muto, T, Tsuchiya, D, Morikawa, K, Jingami, H. | | Deposit date: | 2006-12-17 | | Release date: | 2007-02-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structures of the extracellular regions of the group II/III metabotropic glutamate receptors
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
1KAK
 
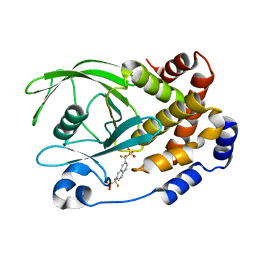 | | Human Tyrosine Phosphatase 1B Complexed with an Inhibitor | | Descriptor: | PROTEIN-TYROSINE PHOSPHATASE, NON-RECEPTOR TYPE 1, {[7-(DIFLUORO-PHOSPHONO-METHYL)-NAPHTHALEN-2-YL]-DIFLUORO-METHYL}-PHOSPHONIC ACID | | Authors: | Jia, Z, Ye, Q, Dinaut, A.N, Wang, Q, Waddleton, D, Payette, P, Ramachandran, C, Kennedy, B, Hum, G, Taylor, S.D. | | Deposit date: | 2001-11-02 | | Release date: | 2002-06-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of protein tyrosine phosphatase 1B in complex with inhibitors bearing two phosphotyrosine mimetics.
J.Med.Chem., 44, 2001
|
|
6MRS
 
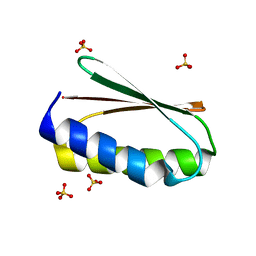 | |
6MSR
 
 | | Crystal structure of pRO-2.5 | | Descriptor: | pRO-2.5 | | Authors: | Bick, M.J, Sankaran, B, Boyken, S.E, Baker, D. | | Deposit date: | 2018-10-17 | | Release date: | 2019-05-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | De novo design of tunable, pH-driven conformational changes.
Science, 364, 2019
|
|
3MHO
 
 | | Crystal structure of human carbonic anhydrase isozyme II with 4-[N-(6-chloro-5-formyl-2-methylthiopyrimidin-4-yl)amino]benzenesulfonamide | | Descriptor: | 4-{[6-chloro-5-formyl-2-(methylsulfanyl)pyrimidin-4-yl]amino}benzenesulfonamide, Carbonic anhydrase 2, DIMETHYL SULFOXIDE, ... | | Authors: | Grazulis, S, Manakova, E, Golovenko, D, Sukackaite, R. | | Deposit date: | 2010-04-08 | | Release date: | 2010-10-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | 4-[N-(Substituted 4-pyrimidinyl)amino]benzenesulfonamides as inhibitors of carbonic anhydrase isozymes I, II, VII, and XIII
Bioorg.Med.Chem., 18, 2010
|
|
6MTT
 
 | | Crystal structure of VRC46.01 Fab in complex with gp41 peptide | | Descriptor: | Antibody VRC46.01 Fab heavy chain, Antibody VRC46.01 Fb light chain, RV217 founder virus gp41 peptide | | Authors: | Kwon, Y.D, Druz, A, Law, W.H, Peng, D, Veradi, R, Doria-Rose, N.A, Kwong, P.D. | | Deposit date: | 2018-10-21 | | Release date: | 2019-03-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Longitudinal Analysis Reveals Early Development of Three MPER-Directed Neutralizing Antibody Lineages from an HIV-1-Infected Individual.
Immunity, 50, 2019
|
|
6MZ0
 
 | | Crystal structure of Tdp1 catalytic domain in complex with compound XZ530 | | Descriptor: | 1,2-ETHANEDIOL, 5-hydroxypyrazine-2,3-dicarboxylic acid, Tyrosyl-DNA phosphodiesterase 1 | | Authors: | Lountos, G.T, Zhao, X.Z, Kiselev, E, Tropea, J.E, Needle, D, Burke Jr, T.R, Pommier, Y, Waugh, D.S. | | Deposit date: | 2018-11-02 | | Release date: | 2019-11-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.969 Å) | | Cite: | Structure of Tdp1 catalytic domain in complex with compound XZ530
To Be Published
|
|
6N0R
 
 | | Crystal structure of Tdp1 catalytic domain in complex with compound XZ572 | | Descriptor: | 1,2-ETHANEDIOL, 4-(methylamino)benzene-1,2-dicarboxylic acid, DIMETHYL SULFOXIDE, ... | | Authors: | Lountos, G.T, Zhao, X.Z, Kiselev, E, Tropea, J.E, Needle, D, Burke Jr, T.R, Pommier, Y, Waugh, D.S. | | Deposit date: | 2018-11-07 | | Release date: | 2019-11-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.544 Å) | | Cite: | Crystal structure of Tdp1 catalytic domain
To Be Published
|
|
7UQC
 
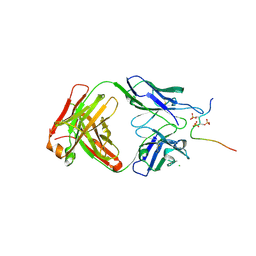 | |
6N50
 
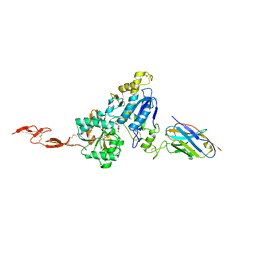 | | Metabotropic Glutamate Receptor 5 Extracellular Domain in Complex with Nb43 and L-quisqualic acid | | Descriptor: | (S)-2-AMINO-3-(3,5-DIOXO-[1,2,4]OXADIAZOLIDIN-2-YL)-PROPIONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, Metabotropic glutamate receptor 5, ... | | Authors: | Koehl, A, Hu, H, Feng, D, Sun, B, Chu, M, Weis, W.I, Skiniotis, G, Mathiesen, J.M, Kobilka, B.K. | | Deposit date: | 2018-11-20 | | Release date: | 2019-01-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.751 Å) | | Cite: | Structural insights into the activation of metabotropic glutamate receptors.
Nature, 566, 2019
|
|
6N38
 
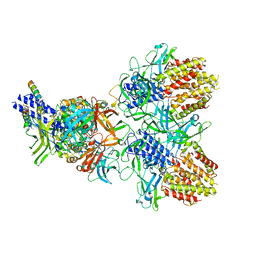 | | Structure of the type VI secretion system TssK-TssF-TssG baseplate subcomplex revealed by cryo-electron microscopy - full map sharpened | | Descriptor: | Putative type VI secretion protein, Unassigned protein | | Authors: | Park, Y.J, Lacourse, K.D, Cambillau, C, Seattle Structural Genomics Center for Infectious Disease (SSGCID), DiMaio, F, Mougous, J.D, Veesler, D. | | Deposit date: | 2018-11-14 | | Release date: | 2018-12-26 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structure of the type VI secretion system TssK-TssF-TssG baseplate subcomplex revealed by cryo-electron microscopy.
Nat Commun, 9, 2018
|
|
1K6Q
 
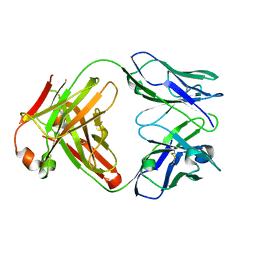 | | Crystal structure of antibody Fab fragment D3 | | Descriptor: | immunoglobulin Fab D3, heavy chain, light chain | | Authors: | Faelber, K, Kelley, R.F, Kirchhofer, D, Muller, Y.A. | | Deposit date: | 2001-10-17 | | Release date: | 2002-04-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The crystal structure of murine Fab D3 at 2.4 A resolution in comparison with the humanised Fab D3h44 (1.85A) provides structural insight into the humanisation process of the D3 anti-tissue factor antibody
To be Published
|
|
2BTI
 
 | | Structure-function studies of the RmsA CsrA post-transcriptional global regulator protein family reveals a class of RNA-binding structure | | Descriptor: | ACETATE ION, CARBON STORAGE REGULATOR HOMOLOG, SULFATE ION | | Authors: | Heeb, S, Kuehne, S.A, Bycroft, M, Crivii, S, Allen, M.D, Haas, D, Camara, M, Williams, P. | | Deposit date: | 2005-05-31 | | Release date: | 2006-01-04 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Functional Analysis of the Post-Transcriptional Regulator Rsma Reveals a Novel RNA-Binding Site.
J.Mol.Biol., 355, 2006
|
|
2BJI
 
 | | High Resolution Structure of myo-Inositol Monophosphatase, The Target of Lithium Therapy | | Descriptor: | INOSITOL-1(OR 4)-MONOPHOSPHATASE, MAGNESIUM ION | | Authors: | Gill, R, Mohammed, F, Badyal, R, Coates, L, Erskine, P, Thompson, D, Cooper, J, Gore, M, Wood, S. | | Deposit date: | 2005-02-03 | | Release date: | 2005-02-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | High-resolution structure of myo-inositol monophosphatase, the putative target of lithium therapy.
Acta Crystallogr. D Biol. Crystallogr., 61, 2005
|
|
3N52
 
 | | crystal Structure analysis of MIP2 | | Descriptor: | C-X-C motif chemokine 2 | | Authors: | Rajasekaran, D. | | Deposit date: | 2010-05-24 | | Release date: | 2011-06-08 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A Model of GAG/MIP-2/CXCR2 Interfaces and Its Functional Effects.
Biochemistry, 51, 2012
|
|
2BJN
 
 | | X-ray Structure of human TPC6 | | Descriptor: | GLYCEROL, SULFATE ION, TRAFFICKING PROTEIN PARTICLE COMPLEX SUBUNIT 6B | | Authors: | Kummel, D, Mueller, J.J, Roske, Y, Misselwitz, R, Bussow, K, Heinemann, U. | | Deposit date: | 2005-02-04 | | Release date: | 2005-07-20 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Structure of the Trapp Subunit Tpc6 Suggests a Model for a Trapp Subcomplex.
Embo Rep., 6, 2005
|
|
6N17
 
 | | Crystal structure of Tdp1 catalytic domain in complex with compound XZ577 | | Descriptor: | 1,2-ETHANEDIOL, 4-[(3-carboxypropanoyl)amino]benzene-1,2-dicarboxylic acid, DIMETHYL SULFOXIDE, ... | | Authors: | Lountos, G.T, Zhao, X.Z, Kiselev, E, Tropea, J.E, Needle, D, Burke Jr, T.R, Pommier, Y, Waugh, D.S. | | Deposit date: | 2018-11-08 | | Release date: | 2019-07-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.639 Å) | | Cite: | Identification of a ligand binding hot spot and structural motifs replicating aspects of tyrosyl-DNA phosphodiesterase I (TDP1) phosphoryl recognition by crystallographic fragment cocktail screening.
Nucleic Acids Res., 47, 2019
|
|
2BKW
 
 | | Yeast alanine:glyoxylate aminotransferase YFL030w | | Descriptor: | ALANINE-GLYOXYLATE AMINOTRANSFERASE 1, GLYOXYLIC ACID, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Meyer, P, Liger, D, Leulliot, N, Quevillon-Cheruel, S, Zhou, C.Z, Borel, F, Ferrer, J.L, Poupon, A, Janin, J, van Tilbeurgh, H. | | Deposit date: | 2005-02-21 | | Release date: | 2005-11-02 | | Last modified: | 2015-11-18 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Crystal Structure and Confirmation of the Alanine:Glyoxylate Aminotransferase Activity of the Yfl030W Yeast Protein
Biochimie, 87, 2005
|
|
2BNK
 
 | | The structure of phage phi29 replication organizer protein p16.7 | | Descriptor: | EARLY PROTEIN GP16.7 | | Authors: | Albert, A, Asensio, J.L, Munoz-Espin, D, Gonzalez, C, Hermoso, J.A, Villar, L, Jimenez-Barbero, J, Salas, M, Meijer, W.J.J. | | Deposit date: | 2005-03-28 | | Release date: | 2005-04-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of the Functional Domain of {Varphi}29 Replication Organizer: Insights Into Oligomerization and DNA Binding.
J.Biol.Chem., 280, 2005
|
|
2BUD
 
 | | The solution structure of the chromo barrel domain from the males- absent on the first (MOF) protein | | Descriptor: | MALES-ABSENT ON THE FIRST PROTEIN | | Authors: | Nielsen, P.R, Nietlispach, D, Buscaino, A, Warner, R.J, Akhtar, A, Murzin, A.G, Murzina, N.V, Laue, E.D. | | Deposit date: | 2005-06-09 | | Release date: | 2005-06-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of the Chromo Barrel Domain from the Mof Acetyltransferase
J.Biol.Chem., 280, 2005
|
|
2BVB
 
 | | The C-terminal domain from Micronemal Protein 1 (MIC1) from Toxoplasma Gondii | | Descriptor: | MICRONEMAL PROTEIN 1 | | Authors: | Saouros, S, Edwards-Jones, B, Reiss, M, Sawmynaden, K, Cota, E, Simpson, P, Dowse, T.J, Jakle, U, Ramboarina, S, Shivarattan, T, Matthews, S, Soldati-Favre, D. | | Deposit date: | 2005-06-23 | | Release date: | 2005-10-12 | | Last modified: | 2021-06-23 | | Method: | SOLUTION NMR | | Cite: | A Novel Galectin-Like Domain from Toxoplasma Gondll Micronemal Protein 1 Assists the Folding, Assembly,and Transport of a Cell-Adhesion Complex.
J.Biol.Chem., 280, 2005
|
|
2C5B
 
 | | X-ray crystal structure of 5'-fluorodeoxyadenosine synthase from Streptomyces cattleya complexed with 2'deoxy-5'deoxy-fluoroadenosine. | | Descriptor: | 5'-FLUORO-2',5'-DIDEOXYADENOSINE, 5'-FLUORO-5'-DEOXYADENOSINE SYNTHASE, METHIONINE | | Authors: | McEwan, A.R, Deng, H, McGlinchey, R.P, Robinson, D.A, O'Hagan, D, Naismith, J.H, Spencer, J. | | Deposit date: | 2005-10-26 | | Release date: | 2007-02-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Substrate Specificity in Enzymatic Fluorination. The Fluorinase from Streptomyces Cattleya Accepts 2'-Deoxyadenosine Substrates.
Org.Biomol.Chem., 4, 2006
|
|
6MWN
 
 | |
2BOK
 
 | | Factor Xa - cation | | Descriptor: | COAGULATION FACTOR X, SODIUM ION, [AMINO (4-{(3AS,4R,8AS,8BR)-1,3-DIOXO-2- [3-(TRIMETHYLAMMONIO) PROPYL]DECAHYDROPYRROLO[3,4-A] PYRROLIZIN-4-YL}PHENYL) METHYLENE]AMMONIUM | | Authors: | Morgenthaler, M, Schaerer, K, Paulini, R, Obst-Sander, U, Banner, D.W, Schlatter, D, Benz, J, Stihle, M, Diederich, F. | | Deposit date: | 2005-04-12 | | Release date: | 2005-06-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Quantification of Cation-Pi Interactions in Protein-Ligand Complexes: Crystal-Structure Analysis of Factor Xa Bound to a Quaternary Ammonium Ion Ligand
Angew.Chem.Int.Ed.Engl., 44, 2005
|
|
