5MWK
 
 | | Glycoside hydrolase BT_0986 | | Descriptor: | BROMIDE ION, CALCIUM ION, Glycoside hydrolase family 2, ... | | Authors: | Basle, A, Ndeh, D, Rogowski, A, Cartmell, A, Luis, A.S, Venditto, I, Labourel, A, Gilbert, H.J. | | Deposit date: | 2017-01-18 | | Release date: | 2017-03-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Complex pectin metabolism by gut bacteria reveals novel catalytic functions.
Nature, 544, 2017
|
|
8SSN
 
 | | Abl kinase in complex with SKI and asciminib | | Descriptor: | 6,7-dimethoxy-N-(4-phenoxyphenyl)quinazolin-4-amine, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Ludewig, H, Kim, C, Kern, D. | | Deposit date: | 2023-05-08 | | Release date: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | A biophysical framework for double-drugging kinases.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8SSO
 
 | | AurA bound to danusertib and inhibiting monobody Mb2 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Aurora kinase A, ... | | Authors: | Ludewig, H, Kim, C, Kern, D. | | Deposit date: | 2023-05-08 | | Release date: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | A biophysical framework for double-drugging kinases.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
1C9W
 
 | | CHO REDUCTASE WITH NADP+ | | Descriptor: | CHO REDUCTASE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Ye, Q, Li, X, Hyndman, D, Flynn, T.G, Jia, Z. | | Deposit date: | 1999-08-03 | | Release date: | 2000-01-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of CHO reductase, a member of the aldo-keto reductase superfamily.
Proteins, 38, 2000
|
|
8SSP
 
 | | AurA bound to danusertib and activating monobody Mb1 | | Descriptor: | 1,2-ETHANEDIOL, 1,3-PROPANDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Ludewig, H, Kim, C, Kern, D. | | Deposit date: | 2023-05-08 | | Release date: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A biophysical framework for double-drugging kinases.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8TAR
 
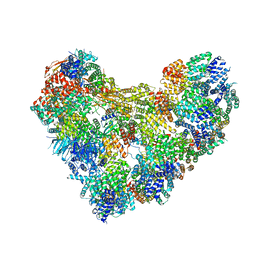 | | APC/C-CDH1-UBE2C-Ubiquitin-CyclinB-NTD | | Descriptor: | Anaphase-promoting complex subunit 1, Anaphase-promoting complex subunit 10, Anaphase-promoting complex subunit 11, ... | | Authors: | Bodrug, T, Welsh, K.A, Bolhuis, D.L, Paulakonis, E, Martinez-Chacin, R.C, Liu, B, Pinkin, N, Bonacci, T, Cui, L, Xu, P, Roscow, O, Amann, S.J, Grishkovskaya, I, Emanuele, M.J, Harrison, J.S, Steimel, J.P, Hahn, K.M, Zhang, W, Zhong, E, Haselbach, D, Brown, N.G. | | Deposit date: | 2023-06-27 | | Release date: | 2023-09-27 | | Last modified: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Time-resolved cryo-EM (TR-EM) analysis of substrate polyubiquitination by the RING E3 anaphase-promoting complex/cyclosome (APC/C).
Nat.Struct.Mol.Biol., 30, 2023
|
|
5MTI
 
 | | Bamb_5917 Acyl-Carrier Protein | | Descriptor: | Phosphopantetheine-binding protein | | Authors: | Gallo, A, Kosol, S, Griffiths, D, Masschelein, J, Alkhalaf, L, Smith, H, Valentic, T, Tsai, S, Challis, G, Lewandowski, J.R. | | Deposit date: | 2017-01-09 | | Release date: | 2018-08-01 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structural basis for chain release from the enacyloxin polyketide synthase
Nat.Chem., 2019
|
|
8T6V
 
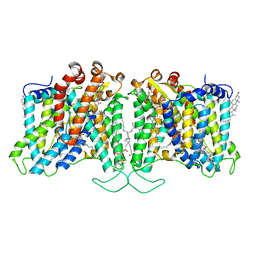 | | Cryo-EM structure of human Anion Exchanger 1 bound to 4,4'-Diisothiocyanatostilbene-2,2'-Disulfonic Acid (DIDS) | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4,4'-Diisothiocyano-2,2'-stilbenedisulfonic acid, ... | | Authors: | Capper, M.J, Zilberg, G, Mathiharan, Y.K, Yang, S, Stone, A.C, Wacker, D. | | Deposit date: | 2023-06-18 | | Release date: | 2023-09-13 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Substrate binding and inhibition of the anion exchanger 1 transporter.
Nat.Struct.Mol.Biol., 30, 2023
|
|
6BY4
 
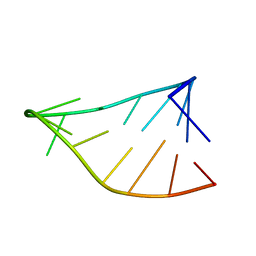 | | Single-State 14-mer UUCG Tetraloop calculated from Exact NOEs | | Descriptor: | RNA (5'-R(P*GP*GP*CP*AP*CP*UP*UP*CP*GP*GP*UP*GP*CP*C)-3') | | Authors: | Nichols, P.J, Henen, M.A, Born, A, Strotz, D, Guntert, P, Vogeli, B. | | Deposit date: | 2017-12-19 | | Release date: | 2018-06-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | High-resolution small RNA structures from exact nuclear Overhauser enhancement measurements without additional restraints.
Commun Biol, 1, 2018
|
|
6BY5
 
 | | Two-State 14-mer UUCG Tetraloop calculated from Exact NOEs (State one: Conformers 1-5, State Two: Conformers 6-10) | | Descriptor: | RNA (5'-R(P*GP*GP*CP*AP*CP*UP*UP*CP*GP*GP*UP*GP*CP*C)-3') | | Authors: | Nichols, P.J, Henen, M.A, Born, A, Strotz, D, Guntert, P, Vogeli, B. | | Deposit date: | 2017-12-19 | | Release date: | 2018-06-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | High-resolution small RNA structures from exact nuclear Overhauser enhancement measurements without additional restraints.
Commun Biol, 1, 2018
|
|
3OY1
 
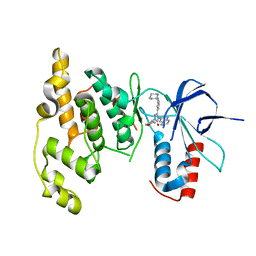 | | Highly Selective c-Jun N-Terminal Kinase (JNK) 2 and 3 Inhibitors with In Vitro CNS-like Pharmacokinetic Properties | | Descriptor: | 5-[2-(cyclohexylamino)pyridin-4-yl]-4-naphthalen-2-yl-2-(tetrahydro-2H-pyran-4-yl)-2,4-dihydro-3H-1,2,4-triazol-3-one, Mitogen-activated protein kinase 10 | | Authors: | Probst, G.D, Bowers, S, Sealy, J.M, Truong, A, Neitz, J, Hom, R.K, Galemmo Jr, R.A, Konradi, A.W, Sham, H.L, Quincy, D, Pan, H, Yao, N. | | Deposit date: | 2010-09-22 | | Release date: | 2011-08-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Highly selective c-Jun N-terminal kinase (JNK) 2 and 3 inhibitors with in vitro CNS-like pharmacokinetic properties prevent neurodegeneration.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
6BNH
 
 | | Solution NMR structures of BRD4 ET domain with JMJD6 peptide | | Descriptor: | Bifunctional arginine demethylase and lysyl-hydroxylase JMJD6, Bromodomain-containing protein 4 | | Authors: | Konuma, T, Yu, D, Zhao, C, Ju, Y, Sharma, R, Ren, C, Zhang, Q, Zhou, M.-M, Zeng, L. | | Deposit date: | 2017-11-16 | | Release date: | 2017-12-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Mechanism of the Oxygenase JMJD6 Recognition by the Extraterminal (ET) Domain of BRD4.
Sci Rep, 7, 2017
|
|
1C94
 
 | | REVERSING THE SEQUENCE OF THE GCN4 LEUCINE ZIPPER DOES NOT AFFECT ITS FOLD. | | Descriptor: | RETRO-GCN4 LEUCINE ZIPPER | | Authors: | Mittl, P.R.E, Deillon, C.A, Sargent, D, Liu, N, Klauser, S, Thomas, R.M, Gutte, B, Gruetter, M.G. | | Deposit date: | 1999-07-30 | | Release date: | 2000-03-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | The retro-GCN4 leucine zipper sequence forms a stable three-dimensional structure.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
8TAU
 
 | | APC/C-CDH1-UBE2C-UBE2S-Ubiquitin-CyclinB | | Descriptor: | Anaphase-promoting complex subunit 1, Anaphase-promoting complex subunit 10, Anaphase-promoting complex subunit 11, ... | | Authors: | Bodrug, T, Welsh, K.A, Bolhuis, D.L, Paulakonis, E, Martinez-Chacin, R.C, Liu, B, Pinkin, N, Bonacci, T, Cui, L, Xu, P, Roscow, O, Amann, S.J, Grishkovskaya, I, Emanuele, M.J, Harrison, J.S, Steimel, J.P, Hahn, K.M, Zhang, W, Zhong, E, Haselbach, D, Brown, N.G. | | Deposit date: | 2023-06-27 | | Release date: | 2023-09-27 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Time-resolved cryo-EM (TR-EM) analysis of substrate polyubiquitination by the RING E3 anaphase-promoting complex/cyclosome (APC/C).
Nat.Struct.Mol.Biol., 30, 2023
|
|
8SMU
 
 | | Integral fusion of the HtaA CR2 domain from Corynebacterium diphtheriae within EGFP | | Descriptor: | CHLORIDE ION, GLYCEROL, HtaACR2 integral fusion within enhanced green fluorescent protein, ... | | Authors: | Mahoney, B.J, Cascio, D, Clubb, R.T. | | Deposit date: | 2023-04-26 | | Release date: | 2023-09-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Development and atomic structure of a new fluorescence-based sensor to probe heme transfer in bacterial pathogens.
J.Inorg.Biochem., 249, 2023
|
|
3WFQ
 
 | | tRNA processing enzyme complex 1 | | Descriptor: | Poly A polymerase, RNA (73-MER) | | Authors: | Yamashita, S, Takeshita, D, Tomita, K. | | Deposit date: | 2013-07-23 | | Release date: | 2014-01-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.619 Å) | | Cite: | Translocation and rotation of tRNA during template-independent RNA polymerization by tRNA nucleotidyltransferase
Structure, 22, 2014
|
|
5MZ8
 
 | | Crystal structure of aldehyde dehydrogenase 21 (ALDH21) from Physcomitrella patens in complex with the reaction product succinate | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Kopecny, D, Vigouroux, A, Briozzo, P, Morera, S. | | Deposit date: | 2017-01-31 | | Release date: | 2017-08-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The ALDH21 gene found in lower plants and some vascular plants codes for a NADP(+) -dependent succinic semialdehyde dehydrogenase.
Plant J., 92, 2017
|
|
6CGY
 
 | | Structure of the Quorum Quenching lactonase from Alicyclobacillus acidoterrestris bound to a phosphate anion | | Descriptor: | 1,2-ETHANEDIOL, Beta-lactamase, COBALT (II) ION, ... | | Authors: | Bergonzi, C, Schwab, M, Naik, T, Daude, D, Chabriere, E, Elias, M. | | Deposit date: | 2018-02-21 | | Release date: | 2018-08-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural and Biochemical Characterization of AaL, a Quorum Quenching Lactonase with Unusual Kinetic Properties.
Sci Rep, 8, 2018
|
|
3WPP
 
 | | Acinetobacter sp. Tol 5 AtaA YDD-DALL3 domains in C-terminal stalk fused to GCN4 adaptors (CstalkC1iii) | | Descriptor: | CHLORIDE ION, Trimeric autotransporter adhesin | | Authors: | Koiwai, K, Hartmann, M.D, Yoshimoto, S, Nur 'Izzah, N, Suzuki, A, Linke, D, Lupas, A.N, Hori, K. | | Deposit date: | 2014-01-15 | | Release date: | 2015-03-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.952 Å) | | Cite: | Structural Basis for Toughness and Flexibility in the C-terminal Passenger Domain of an Acinetobacter Trimeric Autotransporter Adhesin.
J.Biol.Chem., 291, 2016
|
|
5N4K
 
 | | N-terminal domain of a human Coronavirus NL63 nucleocapsid protein | | Descriptor: | Nucleoprotein, SULFATE ION | | Authors: | Zdzalik, M, Szelazek, B, Kabala, W, Golik, P, Burmistrz, M, Florek, D, Kus, K, Pyrc, K, Dubin, G. | | Deposit date: | 2017-02-10 | | Release date: | 2017-02-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Structural Characterization of Human Coronavirus NL63 N Protein.
J. Virol., 91, 2017
|
|
8T6U
 
 | | Cryo-EM structure of human Anion Exchanger 1 bound to Dipyridamole | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-[[2-[bis(2-hydroxyethyl)amino]-4,8-di(piperidin-1-yl)pyrimido[5,4-d]pyrimidin-6-yl]-(2-hydroxyethyl)amino]ethanol, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Capper, M.J, Zilberg, G, Mathiharan, Y.K, Yang, S, Stone, A.C, Wacker, D. | | Deposit date: | 2023-06-18 | | Release date: | 2023-09-13 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Substrate binding and inhibition of the anion exchanger 1 transporter.
Nat.Struct.Mol.Biol., 30, 2023
|
|
1C9O
 
 | | CRYSTAL STRUCTURE ANALYSIS OF THE BACILLUS CALDOLYTICUS COLD SHOCK PROTEIN BC-CSP | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, COLD-SHOCK PROTEIN, SODIUM ION | | Authors: | Mueller, U, Perl, D, Schmid, F.X, Heinemann, U. | | Deposit date: | 1999-08-03 | | Release date: | 2000-04-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Thermal stability and atomic-resolution crystal structure of the Bacillus caldolyticus cold shock protein.
J.Mol.Biol., 297, 2000
|
|
5MZ5
 
 | | Crystal structure of aldehyde dehydrogenase 21 (ALDH21) from Physcomitrella patens in its apoform | | Descriptor: | 1,2-ETHANEDIOL, ALDH21), DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kopecny, D, Koncitikova, R, Briozzo, P, Morera, S. | | Deposit date: | 2017-01-30 | | Release date: | 2017-08-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The ALDH21 gene found in lower plants and some vascular plants codes for a NADP(+) -dependent succinic semialdehyde dehydrogenase.
Plant J., 92, 2017
|
|
3WMU
 
 | | The structure of an anti-cancer lectin mytilec apo-form from the mussel Mytilus galloprovincialis | | Descriptor: | GLYCEROL, Lectin | | Authors: | Terada, D, Kawai, F, Noguchi, H, Unzai, S, Park, S.-Y, Ozeki, Y, Tame, J.R.H. | | Deposit date: | 2013-11-27 | | Release date: | 2014-12-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Crystal structure of MytiLec, a galactose-binding lectin from the mussel Mytilus galloprovincialis with cytotoxicity against certain cancer cell types
Sci Rep, 6, 2016
|
|
3WPM
 
 | | Crystal structure of the anaerobic DesB-gallate complex by co-crystallization | | Descriptor: | 3,4,5-trihydroxybenzoic acid, FE (III) ION, Gallate dioxygenase | | Authors: | Sugimoto, K, Senda, M, Kasai, D, Fukuda, M, Masai, E, Senda, T. | | Deposit date: | 2014-01-14 | | Release date: | 2014-04-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular Mechanism of Strict Substrate Specificity of an Extradiol Dioxygenase, DesB, Derived from Sphingobium sp. SYK-6
Plos One, 9, 2014
|
|
