5HS3
 
 | | Human thymidylate synthase complexed with dUMP and 3-amino-2-benzoyl-4-methylthieno[2,3-b]pyridin-6-ol | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, 3-amino-2-benzoyl-4-methylthieno[2,3-b]pyridin-6-ol, Thymidylate synthase | | Authors: | Chen, D, Almqvist, H, Axelsson, H, Jafari, R, Mateus, A, Haraldsson, M, Larsson, A, Artursson, P, Molina, D.M, Lundback, T, Nordlund, P. | | Deposit date: | 2016-01-25 | | Release date: | 2016-02-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.103 Å) | | Cite: | CETSA screening identifies known and novel thymidylate synthase inhibitors and slow intracellular activation of 5-fluorouracil
Nat Commun, 7, 2016
|
|
4L0C
 
 | | Crystal structure of the N-Fopmylmaleamic acid deformylase Nfo(S94A) from Pseudomonas putida S16 | | Descriptor: | ACETIC ACID, DI(HYDROXYETHYL)ETHER, Deformylase | | Authors: | Chen, D, Lu, Y, Zhang, Z, Wu, G, Xu, P. | | Deposit date: | 2013-05-31 | | Release date: | 2014-06-04 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of the N-Fopmylmaleamic acid deformylase Nfo from Pseudomonas putida S16
To be Published
|
|
6J14
 
 | | Complex structure of GY-14 and PD-1 | | Descriptor: | GY-14 heavy chain V fragment, GY-14 light chain V fragment, Programmed cell death protein 1 | | Authors: | Chen, D, Tan, S, Whang, H, Zhang, H, Chai, Y, Qi, J, Yan, J, Gao, G.F. | | Deposit date: | 2018-12-27 | | Release date: | 2019-11-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The FG Loop of PD-1 Serves as a "Hotspot" for Therapeutic Monoclonal Antibodies in Tumor Immune Checkpoint Therapy.
Iscience, 14, 2019
|
|
6J15
 
 | | Complex structure of GY-5 Fab and PD-1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, GY-5 heavy chain Fab, ... | | Authors: | Chen, D, Tan, S, Zhang, H, Wang, H, Chai, Y, Qi, J, Yan, J, Gao, G.F. | | Deposit date: | 2018-12-27 | | Release date: | 2019-11-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The FG Loop of PD-1 Serves as a "Hotspot" for Therapeutic Monoclonal Antibodies in Tumor Immune Checkpoint Therapy.
Iscience, 14, 2019
|
|
7K3D
 
 | | The structure of NTMT1 in complex with compound DC1-13 | | Descriptor: | N-terminal Xaa-Pro-Lys N-methyltransferase 1, N~2~-{(2S)-1-[(naphthalen-1-yl)acetyl]-2,5-dihydro-1H-pyrrole-2-carbonyl}-L-lysyl-L-argininamide, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Chen, D, Huang, R, Noinaj, N. | | Deposit date: | 2020-09-11 | | Release date: | 2021-04-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Structure-based Discovery of Cell-Potent Peptidomimetic Inhibitors for Protein N-Terminal Methyltransferase 1.
Acs Med.Chem.Lett., 12, 2021
|
|
5X5A
 
 | | Human thymidylate synthase bound with phosphate ion | | Descriptor: | PHOSPHATE ION, Thymidylate synthase | | Authors: | Chen, D, Jansson, A, Larsson, A, Nordlund, P. | | Deposit date: | 2017-02-15 | | Release date: | 2017-06-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structural analyses of human thymidylate synthase reveal a site that may control conformational switching between active and inactive states
J. Biol. Chem., 292, 2017
|
|
5X66
 
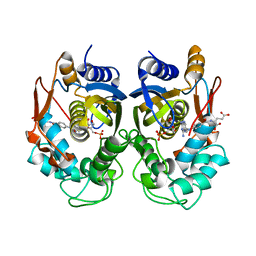 | | Human thymidylate synthase in complex with dUMP and methotrexate | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, METHOTREXATE, Thymidylate synthase | | Authors: | Chen, D, Jansson, A, Larsson, A, Nordlund, P. | | Deposit date: | 2017-02-21 | | Release date: | 2017-06-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural analyses of human thymidylate synthase reveal a site that may control conformational switching between active and inactive states
J. Biol. Chem., 292, 2017
|
|
5X4Y
 
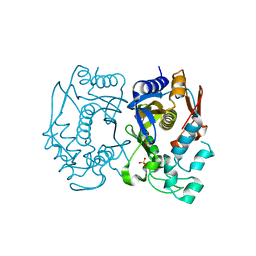 | | Mutant human thymidylate synthase M190K crystallized in a sulfate-containing condition | | Descriptor: | SULFATE ION, Thymidylate synthase | | Authors: | Chen, D, Jansson, A, Larsson, A, Nordlund, P. | | Deposit date: | 2017-02-14 | | Release date: | 2017-06-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural analyses of human thymidylate synthase reveal a site that may control conformational switching between active and inactive states
J. Biol. Chem., 292, 2017
|
|
5X5Q
 
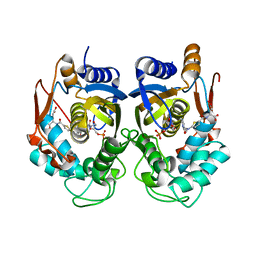 | | Human thymidylate synthase complexed with dUMP and raltitrexed | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, TOMUDEX, Thymidylate synthase | | Authors: | Chen, D, Jansson, A, Larsson, A, Nordlund, P. | | Deposit date: | 2017-02-17 | | Release date: | 2017-06-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Structural analyses of human thymidylate synthase reveal a site that may control conformational switching between active and inactive states
J. Biol. Chem., 292, 2017
|
|
5WRN
 
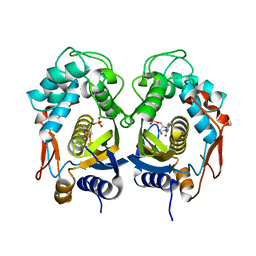 | |
5X69
 
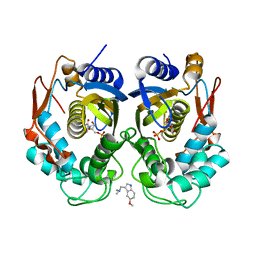 | | Human thymidylate synthase with a fragment bound in the dimer interface | | Descriptor: | 1-(5-methoxy-1H-indol-3-yl)-N,N-dimethyl-methanamine, 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, Thymidylate synthase | | Authors: | Chen, D, Jansson, A, Larsson, A, Nordlund, P. | | Deposit date: | 2017-02-21 | | Release date: | 2017-06-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Structural analyses of human thymidylate synthase reveal a site that may control conformational switching between active and inactive states
J. Biol. Chem., 292, 2017
|
|
5X4W
 
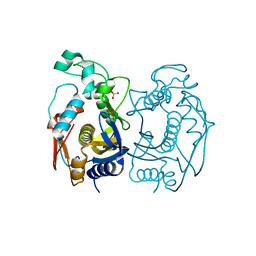 | | Mutant human thymidylate synthase A191K crystallized in a sulfate-free condition | | Descriptor: | PHOSPHATE ION, Thymidylate synthase | | Authors: | Chen, D, Jansson, A, Larsson, A, Nordlund, P. | | Deposit date: | 2017-02-14 | | Release date: | 2017-06-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural analyses of human thymidylate synthase reveal a site that may control conformational switching between active and inactive states
J. Biol. Chem., 292, 2017
|
|
5X5D
 
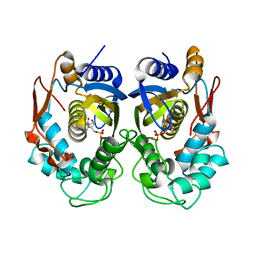 | | Human thymidylate synthase bound with dUMP | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, Thymidylate synthase | | Authors: | Chen, D, Jansson, A, Larsson, A, Nordlund, P. | | Deposit date: | 2017-02-15 | | Release date: | 2017-06-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural analyses of human thymidylate synthase reveal a site that may control conformational switching between active and inactive states
J. Biol. Chem., 292, 2017
|
|
5X67
 
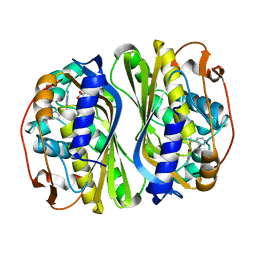 | | Human thymidylate synthase in complex with dUMP and nolatrexed | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, 2-azanyl-6-methyl-5-pyridin-4-ylsulfanyl-3H-quinazolin-4-one, Thymidylate synthase | | Authors: | Chen, D, Jansson, A, Larsson, A, Nordlund, P. | | Deposit date: | 2017-02-21 | | Release date: | 2017-06-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Structural analyses of human thymidylate synthase reveal a site that may control conformational switching between active and inactive states.
J. Biol. Chem., 292, 2017
|
|
5X4X
 
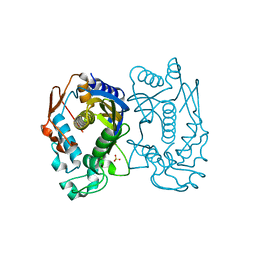 | | Mutant human thymidylate synthase A191K crystallized in a sulfate-containing condition | | Descriptor: | SULFATE ION, Thymidylate synthase | | Authors: | Chen, D, Jansson, A, Larsson, A, Nordlund, P. | | Deposit date: | 2017-02-14 | | Release date: | 2017-06-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structural analyses of human thymidylate synthase reveal a site that may control conformational switching between active and inactive states
J. Biol. Chem., 292, 2017
|
|
5ZNI
 
 | | Plasmodium falciparum purine nucleoside phosphorylase in complex with mefloquine | | Descriptor: | (11R,12S)- Mefloquine, PHOSPHATE ION, Purine nucleoside phosphorylase | | Authors: | Chen, D, Nordlund, P, Dziekan, J.M. | | Deposit date: | 2018-04-09 | | Release date: | 2019-01-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Identifying purine nucleoside phosphorylase as the target of quinine using cellular thermal shift assay.
Sci Transl Med, 11, 2019
|
|
5ZNC
 
 | |
6WJ7
 
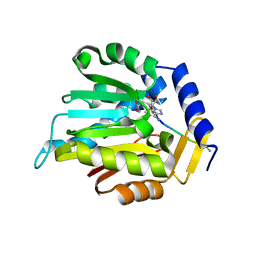 | | The structure of NTMT1 in complex with compound C2A | | Descriptor: | 5'-{[(3S)-3-amino-3-carboxypropyl](ethyl)amino}-5'-deoxyadenosine, GLY-PRO-LYS-ARG-ILE-ALA-NH2, N-terminal Xaa-Pro-Lys N-methyltransferase 1 | | Authors: | Srinivasan, K, Chen, D, Huang, R, Noinaj, N. | | Deposit date: | 2020-04-13 | | Release date: | 2020-08-19 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Probing the Plasticity in the Active Site of Protein N-terminal Methyltransferase 1 Using Bisubstrate Analogues.
J.Med.Chem., 63, 2020
|
|
6PVB
 
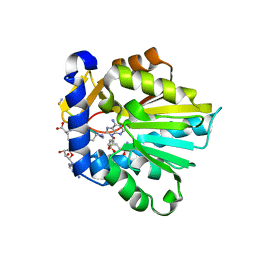 | | The structure of NTMT1 in complex with compound 6 | | Descriptor: | AMINO GROUP-()-(2~{S})-2-azanylpropanal-()-ISOLEUCINE-()-ARGININE-()-LYSINE-()-PROLINE-()-AMINO-ACETALDEHYDE-()-9-(5-{[(3S)-3-amino-3-carboxypropyl](pentyl)amino}-5-deoxy-beta-L-arabinofuranosyl)-9H-purin-6-amine, N-terminal Xaa-Pro-Lys N-methyltransferase 1, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Noinaj, N, Chen, D, Huang, R. | | Deposit date: | 2019-07-20 | | Release date: | 2020-08-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Probing the Plasticity in the Active Site of Protein N-terminal Methyltransferase 1 Using Bisubstrate Analogues.
J.Med.Chem., 63, 2020
|
|
6PVS
 
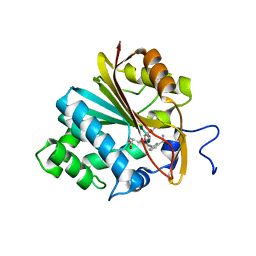 | | Structure of Nicotinamide N-Methyltransferase (NNMT) in complex with inhibitor LL320 | | Descriptor: | 9-(5-{[(3R)-3-amino-3-carboxypropyl][3-(3-carbamoylphenyl)prop-2-yn-1-yl]amino}-5-deoxy-alpha-D-lyxofuranosyl)-9H-purin-6-amine, NNMT protein | | Authors: | Noinaj, N, Huang, R, Chen, D, Yadav, R. | | Deposit date: | 2019-07-21 | | Release date: | 2019-11-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.575 Å) | | Cite: | Novel Propargyl-Linked Bisubstrate Analogues as Tight-Binding Inhibitors for NicotinamideN-Methyltransferase.
J.Med.Chem., 62, 2019
|
|
6PVA
 
 | | The structure of NTMT1 in complex with compound 11 | | Descriptor: | AMINO GROUP-()-LYSINE-()-LYSINE-()-PROLINE-()-AMINO-ACETALDEHYDE-()-5'-{[(3S)-3-amino-3-carboxypropyl](3-aminopropyl)amino}-5'-deoxyadenosine, N-terminal Xaa-Pro-Lys N-methyltransferase 1 | | Authors: | Noinaj, N, Chen, D, Huang, R. | | Deposit date: | 2019-07-20 | | Release date: | 2020-08-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | The structure of NTMT1 in complex with compound 11
To Be published
|
|
6PVE
 
 | | Structure of Nicotinamide N-Methyltransferase (NNMT) in complex with inhibitor LL319 | | Descriptor: | 9-(5-{[(3S)-3-amino-3-carboxypropyl][3-(3-carbamoylphenyl)propyl]amino}-5-deoxy-alpha-D-ribofuranosyl)-9H-purin-6-amine, NNMT protein | | Authors: | Noinaj, N, Huang, R, Chen, D, Yadav, R. | | Deposit date: | 2019-07-20 | | Release date: | 2019-11-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Novel Propargyl-Linked Bisubstrate Analogues as Tight-Binding Inhibitors for NicotinamideN-Methyltransferase.
J.Med.Chem., 62, 2019
|
|
9CUO
 
 | | Crystal structure of CRBN with compound 3 | | Descriptor: | (3S)-3-(3-methyl-2-oxo-2,3-dihydro-1H-1,3-benzimidazol-1-yl)piperidine-2,6-dione, 1,2-ETHANEDIOL, Protein cereblon, ... | | Authors: | Zheng, X, Ji, N, Campbell, V, Slavin, A, Zhu, X, Chen, D, Rong, H, Enerson, B, Mayo, M, Sharma, K, Browne, C.M, Klaus, C.R, Li, H, Massa, G, McDonald, A.A, Shi, Y, Sintchak, M, Skouras, S, Walther, D.M, Yuan, K, Zhang, Y, Kelleher, J, Guang, L, Luo, X, Mainolfi, N, Weiss, M.M. | | Deposit date: | 2024-07-26 | | Release date: | 2024-08-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery of KT-474─a Potent, Selective, and Orally Bioavailable IRAK4 Degrader for the Treatment of Autoimmune Diseases.
J.Med.Chem., 67, 2024
|
|
6K05
 
 | | Crystal structure of BRD2(BD1)with ligand BY27 bound | | Descriptor: | (6~{R})-~{N}-(4-chlorophenyl)-1-methyl-8-(1-methylpyrazol-4-yl)-5,6-dihydro-4~{H}-[1,2,4]triazolo[4,3-a][1]benzazepin-6-amine, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Bromodomain-containing protein 2 | | Authors: | Lu, T, Lu, W, Chen, D, Zhao, Y, Luo, C. | | Deposit date: | 2019-05-05 | | Release date: | 2019-09-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.935 Å) | | Cite: | Discovery, structural insight, and bioactivities of BY27 as a selective inhibitor of the second bromodomains of BET proteins.
Eur.J.Med.Chem., 182, 2019
|
|
6K04
 
 | | Crystal structure of BRD2(BD2)with ligand BY27 bound | | Descriptor: | (6~{R})-~{N}-(4-chlorophenyl)-1-methyl-8-(1-methylpyrazol-4-yl)-5,6-dihydro-4~{H}-[1,2,4]triazolo[4,3-a][1]benzazepin-6-amine, Bromodomain-containing protein 2 | | Authors: | Lu, T, Lu, W, Chen, D, Zhao, Y, Luo, C. | | Deposit date: | 2019-05-05 | | Release date: | 2019-09-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.251 Å) | | Cite: | Discovery, structural insight, and bioactivities of BY27 as a selective inhibitor of the second bromodomains of BET proteins.
Eur.J.Med.Chem., 182, 2019
|
|
