4OBE
 
 | | Crystal Structure of GDP-bound Human KRas | | 分子名称: | GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | 著者 | Hunter, J.C, Gurbani, D, Chen, Z, Westover, K.D. | | 登録日 | 2014-01-07 | | 公開日 | 2014-06-04 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.24 Å) | | 主引用文献 | In situ selectivity profiling and crystal structure of SML-8-73-1, an active site inhibitor of oncogenic K-Ras G12C.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3QDZ
 
 | | Crystal structure of the human thrombin mutant D102N in complex with the extracellular fragment of human PAR4. | | 分子名称: | Proteinase-activated receptor 4, Thrombin heavy chain, Thrombin light chain | | 著者 | Gandhi, P, Chen, Z, Appelbaum, E, Zapata, F, Di Cera, E. | | 登録日 | 2011-01-19 | | 公開日 | 2011-06-29 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structural basis of thrombin-protease-receptor interactions
IUBMB LIFE, 63, 2011
|
|
3QGN
 
 | | The allosteric E*-E equilibrium is a key property of the trypsin fold | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, IODIDE ION, ... | | 著者 | Niu, W, Gohara, D, Chen, Z, Di Cera, E. | | 登録日 | 2011-01-24 | | 公開日 | 2011-07-06 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Crystallographic and kinetic evidence of allostery in a trypsin-like protease.
Biochemistry, 50, 2011
|
|
2KAI
 
 | | REFINED 2.5 ANGSTROMS X-RAY CRYSTAL STRUCTURE OF THE COMPLEX FORMED BY PORCINE KALLIKREIN A AND THE BOVINE PANCREATIC TRYPSIN INHIBITOR. CRYSTALLIZATION, PATTERSON SEARCH, STRUCTURE DETERMINATION, REFINEMENT, STRUCTURE AND COMPARISON WITH ITS COMPONENTS AND WITH THE BOVINE TRYPSIN-PANCREATIC TRYPSIN INHIBITOR COMPLEX | | 分子名称: | BOVINE PANCREATIC TRYPSIN INHIBITOR, KALLIKREIN A | | 著者 | Bode, W, Chen, Z. | | 登録日 | 1984-05-21 | | 公開日 | 1984-07-19 | | 最終更新日 | 2024-06-05 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Refined 2.5 A X-ray crystal structure of the complex formed by porcine kallikrein A and the bovine pancreatic trypsin inhibitor. Crystallization, Patterson search, structure determination, refinement, structure and comparison with its components and with the bovine trypsin-pancreatic trypsin inhibitor complex
J.Mol.Biol., 164, 1983
|
|
2L68
 
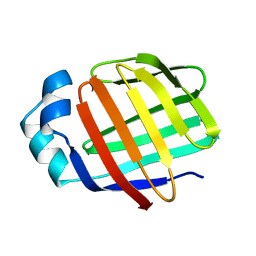 | | Solution Structure of Human Holo L-FABP | | 分子名称: | Fatty acid-binding protein, liver | | 著者 | Cai, J, Luecke, C, Chen, Z, Qiao, Y, Klimtchuk, E.S, Hamilton, J.A. | | 登録日 | 2010-11-17 | | 公開日 | 2011-11-23 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure and backbone dynamics of human liver fatty acid binding protein: fatty acid binding revisited.
Biophys.J., 102, 2012
|
|
2L67
 
 | | Solution Structure of Human Apo L-FABP | | 分子名称: | Fatty acid-binding protein, liver | | 著者 | Cai, J, Luecke, C, Chen, Z, Qiao, Y, Klimtchuk, E.S, Hamilton, J.A. | | 登録日 | 2010-11-17 | | 公開日 | 2011-11-23 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure and backbone dynamics of human liver fatty acid binding protein: fatty acid binding revisited.
Biophys.J., 102, 2012
|
|
2MLD
 
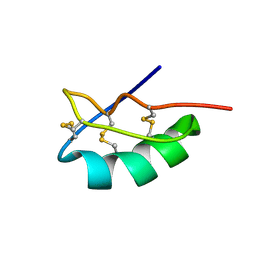 | |
2H47
 
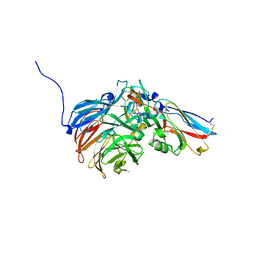 | | Crystal Structure of an Electron Transfer Complex Between Aromatic Amine Dephydrogenase and Azurin from Alcaligenes Faecalis (Form 1) | | 分子名称: | Aromatic Amine Dehydrogenase, Azurin, COPPER (II) ION | | 著者 | Sukumar, N, Chen, Z, Leys, D, Scrutton, N.S, Ferrati, D, Merli, A, Rossi, G.L, Bellamy, H.D, Chistoserdov, A, Davidson, V.L, Mathews, F.S. | | 登録日 | 2006-05-23 | | 公開日 | 2006-11-21 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Crystal Structure of an Electron Transfer Complex between Aromatic Amine Dehydrogenase and Azurin from Alcaligenes faecalis.
Biochemistry, 45, 2006
|
|
2R9E
 
 | |
2RAC
 
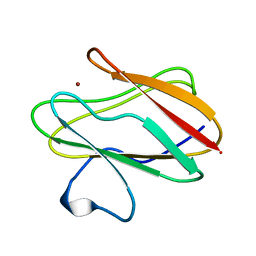 | | AMICYANIN REDUCED, PH 7.7, 1.3 ANGSTROMS | | 分子名称: | COPPER (I) ION, PROTEIN (AMICYANIN) | | 著者 | Cunane, L.M, Chen, Z.-W, Durley, R.C.E, Mathews, F.S. | | 登録日 | 1998-10-02 | | 公開日 | 1998-10-07 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Molecular basis for interprotein complex-dependent effects on the redox properties of amicyanin.
Biochemistry, 37, 1998
|
|
8W2F
 
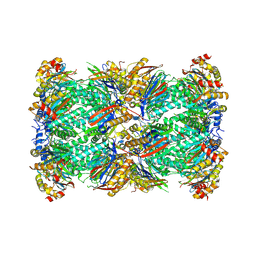 | | Plasmodium falciparum 20S proteasome bound to an inhibitor | | 分子名称: | (3S)-1-[(2-fluoroethoxy)acetyl]-N-{[(4P)-4-(6-methylpyridin-3-yl)-1,3-thiazol-2-yl]methyl}piperidine-3-carboxamide, Proteasome endopeptidase complex, Proteasome subunit alpha type, ... | | 著者 | Han, Y, Deng, X, Ray, S, Chen, Z, Phillips, M. | | 登録日 | 2024-02-20 | | 公開日 | 2024-07-31 | | 最終更新日 | 2024-11-13 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Identification of potent and reversible piperidine carboxamides that are species-selective orally active proteasome inhibitors to treat malaria.
Cell Chem Biol, 31, 2024
|
|
7W8L
 
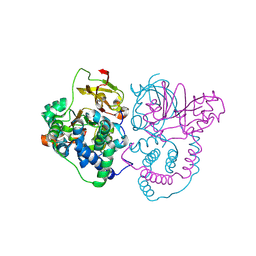 | | Crystal Structure of Co-type nitrile hydratase mutant from Pseudonocardia thermophila - M46R | | 分子名称: | COBALT (II) ION, Cobalt-containing nitrile hydratase subunit beta, Nitrile hydratase | | 著者 | Ma, D, Cheng, Z.Y, Hou, X.D, Peplowski, L, Lai, Q.P, Fu, K, Yin, D.J, Rao, Y.J, Zhou, Z.M. | | 登録日 | 2021-12-08 | | 公開日 | 2022-08-24 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (2.301 Å) | | 主引用文献 | Insight into the broadened substrate scope of nitrile hydratase by static and dynamic structure analysis.
Chem Sci, 13, 2022
|
|
7W8M
 
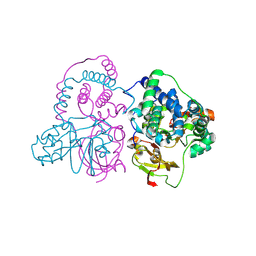 | | Crystal structure of Co-type nitrile hydratase mutant from Pseudomonas thermophila - A129R | | 分子名称: | COBALT (II) ION, Cobalt-containing nitrile hydratase subunit beta, Nitrile hydratase | | 著者 | Ma, D, Cheng, Z.Y, Hou, X.D, Peplowski, L, Lai, Q.P, Fu, K, Yin, D.J, Rao, Y.J, Zhou, Z.M. | | 登録日 | 2021-12-08 | | 公開日 | 2022-08-24 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Insight into the broadened substrate scope of nitrile hydratase by static and dynamic structure analysis.
Chem Sci, 13, 2022
|
|
2M01
 
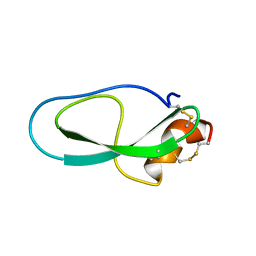 | | Solution structure of Kunitz-type neurotoxin LmKKT-1a from scorpion venom | | 分子名称: | Protease inhibitor LmKTT-1a | | 著者 | Luo, F, Jiang, L, Liu, M, Chen, Z, Wu, Y. | | 登録日 | 2012-10-15 | | 公開日 | 2013-11-13 | | 最終更新日 | 2024-10-30 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Genomic and structural characterization of Kunitz-type peptide LmKTT-1a highlights diversity and evolution of scorpion potassium channel toxins.
Plos One, 8, 2013
|
|
7C2J
 
 | | Crystal structure of nsp16-nsp10 heterodimer from SARS-CoV-2 in complex with SAM (without additional SAM during crystallization) | | 分子名称: | 2'-O-methyltransferase, Non-structural protein 10, S-ADENOSYLMETHIONINE, ... | | 著者 | Lin, S, Chen, H, Ye, F, Chen, Z.M, Yang, F.L, Zheng, Y, Cao, Y, Qiao, J.X, Yang, S.Y, Lu, G.W. | | 登録日 | 2020-05-07 | | 公開日 | 2020-05-27 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.799 Å) | | 主引用文献 | Crystal structure of SARS-CoV-2 nsp10/nsp16 2'-O-methylase and its implication on antiviral drug design.
Signal Transduct Target Ther, 5, 2020
|
|
1JXP
 
 | | BK STRAIN HEPATITIS C VIRUS (HCV) NS3-NS4A | | 分子名称: | NS3 SERINE PROTEASE, NS4A, ZINC ION | | 著者 | Yan, Y, Munshi, S, Chen, Z. | | 登録日 | 1997-08-21 | | 公開日 | 1998-01-14 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Complex of NS3 protease and NS4A peptide of BK strain hepatitis C virus: a 2.2 A resolution structure in a hexagonal crystal form.
Protein Sci., 7, 1998
|
|
4FTB
 
 | | Crystal structure of the authentic Flock House virus particle | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, CHLORIDE ION, ... | | 著者 | Speir, J.A, Chen, Z, Reddy, V.S, Johnson, J.E. | | 登録日 | 2012-06-27 | | 公開日 | 2012-08-01 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Structural study of virus assembly intermediates reveals maturation event sequence and a staging position for externalized lytic peptides
to be published, 2012
|
|
6A58
 
 | | Structure of histone demethylase REF6 | | 分子名称: | Lysine-specific demethylase REF6, ZINC ION | | 著者 | Tian, Z, Chen, Z. | | 登録日 | 2018-06-22 | | 公開日 | 2019-06-26 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.57 Å) | | 主引用文献 | Crystal structures of REF6 and its complex with DNA reveal diverse recognition mechanisms.
Cell Discov, 6, 2020
|
|
7C2I
 
 | | Crystal structure of nsp16-nsp10 heterodimer from SARS-CoV-2 in complex with SAM (with additional SAM during crystallization) | | 分子名称: | 2'-O-methyltransferase, Non-structural protein 10, S-ADENOSYLMETHIONINE, ... | | 著者 | Lin, S, Chen, H, Ye, F, Chen, Z.M, Yang, F.L, Zheng, Y, Cao, Y, Qiao, J.X, Yang, S.Y, Lu, G.W. | | 登録日 | 2020-05-07 | | 公開日 | 2020-05-27 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal structure of SARS-CoV-2 nsp10/nsp16 2'-O-methylase and its implication on antiviral drug design.
Signal Transduct Target Ther, 5, 2020
|
|
6TN8
 
 | | Crystal structure of the ACVR1 (ALK2) kinase in complex with the compound BI-9564 | | 分子名称: | 1,2-ETHANEDIOL, 1,4-DIETHYLENE DIOXIDE, 4-[4-[(dimethylamino)methyl]-2,5-dimethoxy-phenyl]-2-methyl-2,7-naphthyridin-1-one, ... | | 著者 | Williams, E.P, Chen, Z, Burgess-Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A.N. | | 登録日 | 2019-12-06 | | 公開日 | 2019-12-18 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.63 Å) | | 主引用文献 | Crystal structure of the ACVR1 (ALK2) kinase in complex with the compound BI-9564
To Be Published
|
|
6A57
 
 | | Structure of histone demethylase REF6 complexed with DNA | | 分子名称: | DNA (5'-D(*CP*TP*TP*TP*CP*TP*CP*TP*GP*TP*TP*TP*TP*GP*TP*C)-3'), DNA (5'-D(*GP*GP*AP*CP*AP*AP*AP*AP*CP*AP*GP*AP*GP*AP*AP*A)-3'), GLYCEROL, ... | | 著者 | Tian, Z, Chen, Z. | | 登録日 | 2018-06-22 | | 公開日 | 2019-06-26 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Crystal structures of REF6 and its complex with DNA reveal diverse recognition mechanisms.
Cell Discov, 6, 2020
|
|
5F17
 
 | |
7DIY
 
 | | Crystal structure of SARS-CoV-2 nsp10 bound to nsp14-exoribonuclease domain | | 分子名称: | MAGNESIUM ION, ZINC ION, nsp10 protein, ... | | 著者 | Lin, S, Chen, H, Chen, Z.M, Yang, F.L, Ye, F, Zheng, Y, Yang, J, Lin, X, Sun, H.L, Wang, L.L, Wen, A, Cao, Y, Lu, G.W. | | 登録日 | 2020-11-19 | | 公開日 | 2021-05-19 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.693 Å) | | 主引用文献 | Crystal structure of SARS-CoV-2 nsp10 bound to nsp14-ExoN domain reveals an exoribonuclease with both structural and functional integrity.
Nucleic Acids Res., 49, 2021
|
|
6L2W
 
 | | Crystal structure of a novel fold protein Gp72 from the freshwater cyanophage Mic1 | | 分子名称: | freshwater cyanophage protein | | 著者 | Wang, Y, Jin, H, Yang, F, Jiang, Y.L, Zhao, Y.Y, Chen, Z.P, Li, W.F, Chen, Y, Zhou, C.Z, Li, Q. | | 登録日 | 2019-10-07 | | 公開日 | 2020-05-13 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.29 Å) | | 主引用文献 | Crystal structure of a novel fold protein Gp72 from the freshwater cyanophage Mic1.
Proteins, 88, 2020
|
|
6A59
 
 | |
