7L73
 
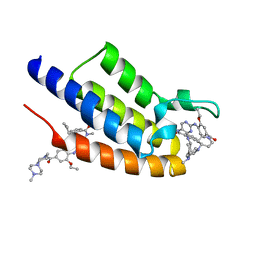 | | Crystal structure of the first bromodomain (BD1) of human BRDT bound to ERK5-IN-1 | | Descriptor: | 11-cyclopentyl-2-({2-ethoxy-4-[4-(4-methylpiperazin-1-yl)piperidine-1-carbonyl]phenyl}amino)-5-methyl-5,11-dihydro-6H-pyrimido[4,5-b][1,4]benzodiazepin-6-one, Bromodomain testis-specific protein | | Authors: | Chan, A, Karim, M.R, Schonbrunn, E. | | Deposit date: | 2020-12-25 | | Release date: | 2021-06-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Differential BET Bromodomain Inhibition by Dihydropteridinone and Pyrimidodiazepinone Kinase Inhibitors.
J.Med.Chem., 64, 2021
|
|
7LEJ
 
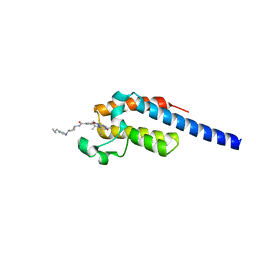 | | Crystal structure of the second bromodomain (BD2) of human BRDT bound to Volasertib | | Descriptor: | Bromodomain testis-specific protein, N-{trans-4-[4-(cyclopropylmethyl)piperazin-1-yl]cyclohexyl}-4-{[(7R)-7-ethyl-5-methyl-8-(1-methylethyl)-6-oxo-5,6,7,8-tetrahydropteridin-2-yl]amino}-3-methoxybenzamide | | Authors: | Chan, A, Karim, M.R, Schonbrunn, E. | | Deposit date: | 2021-01-14 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Differential BET Bromodomain Inhibition by Dihydropteridinone and Pyrimidodiazepinone Kinase Inhibitors.
J.Med.Chem., 64, 2021
|
|
7LEL
 
 | |
7LEM
 
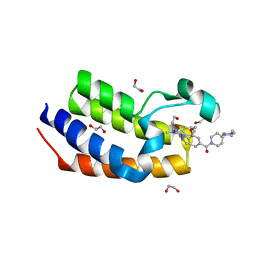 | | Crystal structure of the first bromodomain (BD1) of human BRDT bound to LRRK2-IN-1 | | Descriptor: | 1,2-ETHANEDIOL, 2-[(2-methoxy-4-{[4-(4-methylpiperazin-1-yl)piperidin-1-yl]carbonyl}phenyl)amino]-5,11-dimethyl-5,11-dihydro-6H-pyrimido[4,5-b][1,4]benzodiazepin-6-one, Bromodomain testis-specific protein | | Authors: | Chan, A, Karim, M.R, Schonbrunn, E. | | Deposit date: | 2021-01-14 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Differential BET Bromodomain Inhibition by Dihydropteridinone and Pyrimidodiazepinone Kinase Inhibitors.
J.Med.Chem., 64, 2021
|
|
7LEK
 
 | | Crystal structure of the second bromodomain (BD2) of human BRDT bound to ERK5-IN-1 | | Descriptor: | 1,2-ETHANEDIOL, 11-cyclopentyl-2-({2-ethoxy-4-[4-(4-methylpiperazin-1-yl)piperidine-1-carbonyl]phenyl}amino)-5-methyl-5,11-dihydro-6H-pyrimido[4,5-b][1,4]benzodiazepin-6-one, Bromodomain testis-specific protein, ... | | Authors: | Chan, A, Karim, M.R, Schonbrunn, E. | | Deposit date: | 2021-01-14 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Differential BET Bromodomain Inhibition by Dihydropteridinone and Pyrimidodiazepinone Kinase Inhibitors.
J.Med.Chem., 64, 2021
|
|
7KDW
 
 | |
7K6S
 
 | | Crystal structure of the bromodomain (BD) of human Bromodomain and PHD finger-containing Transcription Factor (BPTF) bound to Compound 4 (SKT1174) | | Descriptor: | (7-amino-3,4-dihydroquinolin-1(2H)-yl)(cyclopropyl)methanone, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Chan, A, Schonbrunn, E. | | Deposit date: | 2020-09-21 | | Release date: | 2020-12-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | New inhibitors for the BPTF bromodomain enabled by structural biology and biophysical assay development.
Org.Biomol.Chem., 18, 2020
|
|
7K6R
 
 | |
7KDZ
 
 | |
5ALB
 
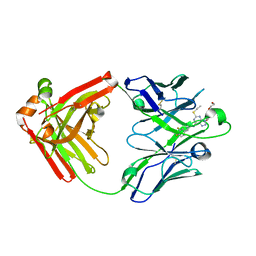 | | Ticagrelor antidote candidate MEDI2452 in complex with ticagrelor | | Descriptor: | MEDI2452 HEAVY CHAIN, MEDI2452 LIGHT CHAIN, Ticagrelor | | Authors: | Buchanan, A, Newton, P, Pehrsson, S, Inghardt, T, Antonsson, T, Svensson, P, Sjogren, T, Oster, L, Janefeldt, A, Sandinge, A, Keyes, F, Austin, M, Spooner, J, Penney, M, Howells, G, Vaughan, T, Nylander, S. | | Deposit date: | 2015-03-07 | | Release date: | 2015-04-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Structural and Functional Characterisation of a Specific Antidote for Ticagrelor.
Blood, 125, 2015
|
|
5ALC
 
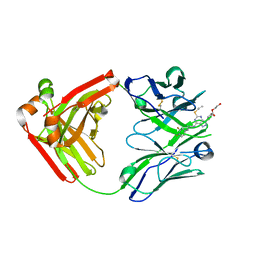 | | Ticagrelor antidote candidate Fab 72 in complex with ticagrelor | | Descriptor: | ANTI-TICAGRELOR FAB 72, HEAVY CHAIN, LIGHT CHAIN, ... | | Authors: | Buchanan, A, Newton, P, Pehrsson, S, Inghardt, T, Antonsson, T, Svensson, P, Sjogren, T, Oster, L, Janefeldt, A, Sandinge, A, Keyes, F, Austin, M, Spooner, J, Penney, M, Howells, G, Vaughan, T, Nylander, S. | | Deposit date: | 2015-03-07 | | Release date: | 2015-04-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and Functional Characterisation of a Specific Antidote for Ticagrelor.
Blood, 125, 2015
|
|
8U9L
 
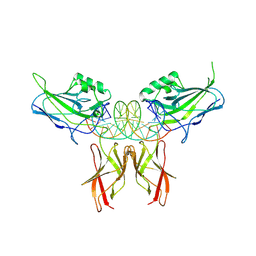 | | Crystal Structure of RelA-cRel chimera complex with DNA | | Descriptor: | DNA (5'-D(P*GP*AP*AP*TP*CP*GP*GP*AP*AP*AP*TP*TP*CP*CP*CP*AP*TP*CP*AP*A)-3'), DNA (5'-D(P*TP*TP*GP*AP*TP*GP*GP*GP*AP*AP*TP*TP*TP*CP*CP*GP*AP*TP*TP*C)-3'), Transcription factor p65,Proto-oncogene c-Rel chimera | | Authors: | Chang, A, Wu, Y, Li, S.X, Smale, S, Chen, L. | | Deposit date: | 2023-09-19 | | Release date: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (3.091 Å) | | Cite: | Crystal Structure of RelA-cRel chimera complex with DNA
To Be Published
|
|
6N81
 
 | |
3IAA
 
 | | Crystal Structure of CalG2, Calicheamicin Glycosyltransferase, TDP bound form | | Descriptor: | CalG2, THYMIDINE-5'-DIPHOSPHATE | | Authors: | Chang, A, Singh, S, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N. | | Deposit date: | 2009-07-13 | | Release date: | 2010-06-02 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.505 Å) | | Cite: | Complete set of glycosyltransferase structures in the calicheamicin biosynthetic pathway reveals the origin of regiospecificity.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3IA7
 
 | | Crystal Structure of CalG4, the Calicheamicin Glycosyltransferase | | Descriptor: | CALCIUM ION, CHLORIDE ION, CalG4 | | Authors: | Chang, A, Singh, S, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N. | | Deposit date: | 2009-07-13 | | Release date: | 2010-06-02 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Complete set of glycosyltransferase structures in the calicheamicin biosynthetic pathway reveals the origin of regiospecificity.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
6N8D
 
 | |
3OTI
 
 | | Crystal Structure of CalG3, Calicheamicin Glycostyltransferase, TDP and calicheamicin T0 bound form | | Descriptor: | CHLORIDE ION, CalG3, Calicheamicin T0, ... | | Authors: | Chang, A, Singh, S, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2010-09-11 | | Release date: | 2010-12-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.597 Å) | | Cite: | Complete set of glycosyltransferase structures in the calicheamicin biosynthetic pathway reveals the origin of regiospecificity.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3OTG
 
 | | Crystal Structure of CalG1, Calicheamicin Glycostyltransferase, TDP bound form | | Descriptor: | CHLORIDE ION, CalG1, THYMIDINE-5'-DIPHOSPHATE | | Authors: | Chang, A, Singh, S, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2010-09-11 | | Release date: | 2010-12-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Complete set of glycosyltransferase structures in the calicheamicin biosynthetic pathway reveals the origin of regiospecificity.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3PKQ
 
 | | Q83D Variant of S. Enterica RmlA with dGTP | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, DI(HYDROXYETHYL)ETHER, Glucose-1-phosphate thymidylyltransferase, ... | | Authors: | Chang, A, Moretti, R, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2010-11-11 | | Release date: | 2011-01-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Expanding the Nucleotide and Sugar 1-Phosphate Promiscuity of Nucleotidyltransferase RmlA via Directed Evolution.
J.Biol.Chem., 286, 2011
|
|
3PKP
 
 | | Q83S Variant of S. Enterica RmlA with dATP | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, Glucose-1-phosphate thymidylyltransferase, MAGNESIUM ION | | Authors: | Chang, A, Moretti, R, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2010-11-11 | | Release date: | 2011-01-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Expanding the Nucleotide and Sugar 1-Phosphate Promiscuity of Nucleotidyltransferase RmlA via Directed Evolution.
J.Biol.Chem., 286, 2011
|
|
3OTH
 
 | | Crystal Structure of CalG1, Calicheamicin Glycostyltransferase, TDP and calicheamicin alpha3I bound form | | Descriptor: | CalG1, Calicheamicin alpha3I, THYMIDINE-5'-DIPHOSPHATE | | Authors: | Chang, A, Singh, S, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2010-09-11 | | Release date: | 2010-12-15 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Complete set of glycosyltransferase structures in the calicheamicin biosynthetic pathway reveals the origin of regiospecificity.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3RSC
 
 | | Crystal Structure of CalG2, Calicheamicin Glycosyltransferase, TDP and calicheamicin T0 bound form | | Descriptor: | CalG2, Calicheamicin T0, PHOSPHATE ION, ... | | Authors: | Chang, A, Helmich, K.E, Singh, S, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2011-05-02 | | Release date: | 2011-08-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Complete set of glycosyltransferase structures in the calicheamicin biosynthetic pathway reveals the origin of regiospecificity.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
8GR3
 
 | |
8GR1
 
 | |
8G9W
 
 | | Cryo-EM structure of vFP49.02 Fab in complex with HIV-1 Env BG505 DS-SOSIP.664 (conformation 1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp120, ... | | Authors: | Changela, A, Gorman, J, Kwong, P.D. | | Deposit date: | 2023-02-22 | | Release date: | 2023-04-19 | | Last modified: | 2023-06-14 | | Method: | ELECTRON MICROSCOPY (4.66 Å) | | Cite: | Diverse Murine Vaccinations Reveal Distinct Antibody Classes to Target Fusion Peptide and Variation in Peptide Length to Improve HIV Neutralization.
J.Virol., 97, 2023
|
|
