7R63
 
 | |
7KGU
 
 | | Structure of 2Q1-Fab, an antibody selective for IDH2R140Q-HLA-B*07:02 | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Miller, M.S, Aytenfisu, T.Y, Wright, K.M, Gabelli, S.B. | | Deposit date: | 2020-10-18 | | Release date: | 2021-06-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural engineering of chimeric antigen receptors targeting HLA-restricted neoantigens.
Nat Commun, 12, 2021
|
|
6B09
 
 | |
7V0C
 
 | | Structure of Ternary Complex of cGAS with dsDNA and Bound 5 -pppG(2 ,5 )pG | | Descriptor: | Cyclic GMP-AMP synthase, MANGANESE (II) ION, Palindromic DNA18, ... | | Authors: | Wu, S, Gabelli, S.B, Sohn, J. | | Deposit date: | 2022-05-10 | | Release date: | 2023-05-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Structure of Ternary Complex of cGAS with dsDNA and Bound 5 -pppG(2 ,5 )pG
To Be Published
|
|
7V0R
 
 | | Structure of Ternary Complex of cGAS with dsDNA and Bound 5 -ppcpG(2 ,5 )pA | | Descriptor: | Cyclic GMP-AMP synthase, MAGNESIUM ION, Palindromic DNA18, ... | | Authors: | Wu, S, Gabelli, S.B, Sohn, J. | | Deposit date: | 2022-05-10 | | Release date: | 2023-05-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structure of Ternary Complex of cGAS with dsDNA and Bound 5 -ppcpG(2 ,5 )pA
To Be Published
|
|
7UTT
 
 | |
5JIN
 
 | | Structure of G9a SET-domain with Histone H3K9M mutant peptide and bound S-adenosylmethionine | | Descriptor: | Histone H3.1 peptide with K9M mutation, Histone-lysine N-methyltransferase EHMT2, S-ADENOSYLMETHIONINE, ... | | Authors: | Jayaram, H, Bellon, S.F, Poy, F. | | Deposit date: | 2016-04-22 | | Release date: | 2016-07-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | S-adenosyl methionine is necessary for inhibition of the methyltransferase G9a by the lysine 9 to methionine mutation on histone H3.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
6UJ8
 
 | | Crystal structure of HLA-B*07:02 with wild-type IDH2 peptide | | Descriptor: | Beta-2-microglobulin, DI(HYDROXYETHYL)ETHER, HLA class I histocompatibility antigen, ... | | Authors: | Miller, M.S, Thirawatananond, P, Gabelli, S.B. | | Deposit date: | 2019-10-02 | | Release date: | 2021-05-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural engineering of chimeric antigen receptors targeting HLA-restricted neoantigens.
Nat Commun, 12, 2021
|
|
6UJ9
 
 | | Crystal structure of HLA-B*07:02 with R140Q mutant IDH2 peptide in complex with Fab | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Beta-2-microglobulin, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Miller, M.S, Thirawatananond, P, Aytenfisu, T.Y, Wright, K, Gabelli, S.B. | | Deposit date: | 2019-10-02 | | Release date: | 2021-05-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural engineering of chimeric antigen receptors targeting HLA-restricted neoantigens.
Nat Commun, 12, 2021
|
|
6UJ7
 
 | | Crystal structure of HLA-B*07:02 with R140Q mutant IDH2 peptide | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, B-7 alpha chain, ... | | Authors: | Miller, M.S, Thirawatananond, P, Gabelli, S.B. | | Deposit date: | 2019-10-02 | | Release date: | 2021-05-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural engineering of chimeric antigen receptors targeting HLA-restricted neoantigens.
Nat Commun, 12, 2021
|
|
5WJA
 
 | | Crystal structure of H107A peptidylglycine alpha-hydroxylating monooxygenase (PHM) in complex with citrate | | Descriptor: | CITRATE ANION, COPPER (II) ION, GLYCEROL, ... | | Authors: | Maheshwari, S, Rudzka, K, Gabelli, S.B, Amzel, L.M. | | Deposit date: | 2017-07-21 | | Release date: | 2018-07-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Effects of copper occupancy on the conformational landscape of peptidylglycine alpha-hydroxylating monooxygenase.
Commun Biol, 1, 2018
|
|
5WKW
 
 | | Crystal structure of apo wild type peptidylglycine alpha-hydroxylating monooxygenase (PHM) | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Peptidyl-glycine alpha-amidating monooxygenase | | Authors: | Maheshwari, S, Rudzka, K, Gabelli, S.B, Amzel, L.M. | | Deposit date: | 2017-07-25 | | Release date: | 2018-07-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Effects of copper occupancy on the conformational landscape of peptidylglycine alpha-hydroxylating monooxygenase.
Commun Biol, 1, 2018
|
|
5WM0
 
 | |
6W51
 
 | |
6NCT
 
 | | Structure of p110alpha/niSH2 - vector data collection | | Descriptor: | Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform, SULFATE ION, ... | | Authors: | Miller, M.S, Maheshwari, S, Amzel, L.M, Gabelli, S.B. | | Deposit date: | 2018-12-12 | | Release date: | 2019-02-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Getting the Most Out of Your Crystals: Data Collection at the New High-Flux, Microfocus MX Beamlines at NSLS-II.
Molecules, 24, 2019
|
|
6NCK
 
 | | Crystal structure of H108A peptidylglycine alpha-hydroxylating monooxygenase (PHM) | | Descriptor: | COPPER (II) ION, NICKEL (II) ION, Peptidyl-glycine alpha-amidating monooxygenase | | Authors: | Miller, M.S, Maheshwari, S, Gabelli, S.B. | | Deposit date: | 2018-12-11 | | Release date: | 2019-02-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Getting the Most Out of Your Crystals: Data Collection at the New High-Flux, Microfocus MX Beamlines at NSLS-II.
Molecules, 24, 2019
|
|
6NCI
 
 | | Crystal structure of CDP-Chase: Vector data collection | | Descriptor: | D-ribose, DI(HYDROXYETHYL)ETHER, PHOSPHATE ION, ... | | Authors: | Miller, M.S, Shi, W, Gabelli, S.B. | | Deposit date: | 2018-12-11 | | Release date: | 2019-02-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Getting the Most Out of Your Crystals: Data Collection at the New High-Flux, Microfocus MX Beamlines at NSLS-II.
Molecules, 24, 2019
|
|
7TAB
 
 | | G-925 bound to the SMARCA4 (BRG1) Bromodomain | | Descriptor: | 2-(6-amino-5-phenylpyridazin-3-yl)phenol, Isoform 4 of Transcription activator BRG1 | | Authors: | Tang, Y, Poy, F, Taylor, A.M, Cochran, A.G, Bellon, S.F. | | Deposit date: | 2021-12-20 | | Release date: | 2022-08-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | GNE-064: A Potent, Selective, and Orally Bioavailable Chemical Probe for the Bromodomains of SMARCA2 and SMARCA4 and the Fifth Bromodomain of PBRM1.
J.Med.Chem., 65, 2022
|
|
7TD9
 
 | | G-059 bound to the SMARCA4 (BRG1) Bromodomain | | Descriptor: | 4-phenyl-5H-pyridazino[4,3-b]indol-3-amine, Isoform 4 of Transcription activator BRG1 | | Authors: | Tang, Y, Poy, F, Taylor, A.M, Cochran, A.G, Bellon, S.F. | | Deposit date: | 2021-12-30 | | Release date: | 2022-08-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | GNE-064: A Potent, Selective, and Orally Bioavailable Chemical Probe for the Bromodomains of SMARCA2 and SMARCA4 and the Fifth Bromodomain of PBRM1.
J.Med.Chem., 65, 2022
|
|
7UYQ
 
 | | Structure of GTP binds to Cyclic GMP AMP synthase (cGAS) through Mg coordination | | Descriptor: | Cyclic GMP-AMP synthase, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Wu, S, Gabelli, S.B, Sohn, J. | | Deposit date: | 2022-05-07 | | Release date: | 2023-05-10 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | The structural basis for 2'-5'/3'-5'-cGAMP synthesis by cGAS
Nat Commun, 15, 2024
|
|
7UYZ
 
 | | Structure of Ternary Complex of cGAS with dsDNA and Bound 5 -pppG(2 ,5 )pG | | Descriptor: | Cyclic GMP-AMP synthase, GUANOSINE-5'-MONOPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Wu, S, Gabelli, S.B, Sohn, J. | | Deposit date: | 2022-05-07 | | Release date: | 2023-05-10 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | The structural basis for 2'-5'/3'-5'-cGAMP synthesis by cGAS
Nat Commun, 15, 2024
|
|
7UZR
 
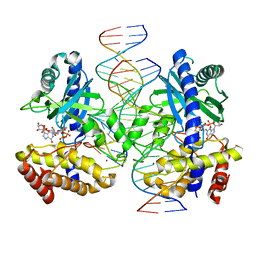 | | Structure of Ternary Complex of cGAS with dsDNA and Bound 5 -pppG(2 ,5 )pG | | Descriptor: | Cyclic GMP-AMP synthase, MANGANESE (II) ION, Palindromic DNA18, ... | | Authors: | Wu, S, Gabelli, S.B, Sohn, J. | | Deposit date: | 2022-05-09 | | Release date: | 2023-05-10 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The structural basis for 2'-5'/3'-5'-cGAMP synthesis by cGAS
Nat Commun, 15, 2024
|
|
5JJ0
 
 | | Structure of G9a SET-domain with Histone H3K9M peptide and excess SAH | | Descriptor: | Histone H3K9M mutant peptide, Histone-lysine N-methyltransferase EHMT2, S-ADENOSYLMETHIONINE, ... | | Authors: | Jayaram, H, Bellon, S.F, Poy, F. | | Deposit date: | 2016-04-22 | | Release date: | 2016-07-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | S-adenosyl methionine is necessary for inhibition of the methyltransferase G9a by the lysine 9 to methionine mutation on histone H3.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5JIY
 
 | | Structure of G9a SET-domain with Histone H3K9norLeucine mutant peptide and bound S-adenosylmethionine | | Descriptor: | Histone H3.1 mutant peptide with H3K9nor-leucine, Histone-lysine N-methyltransferase EHMT2, S-ADENOSYLMETHIONINE, ... | | Authors: | Jayaram, H, Bellon, S.F, Poy, F. | | Deposit date: | 2016-04-22 | | Release date: | 2016-09-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | S-adenosyl methionine is necessary for inhibition of the methyltransferase G9a by the lysine 9 to methionine mutation on histone H3.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
7UXW
 
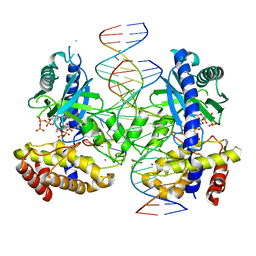 | | Structure of ATP and GTP bind to Cyclic GMP AMP synthase (cGAS) through Mg coordination | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cyclic GMP-AMP synthase, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Wu, S, Gabelli, S.B, Sohn, J. | | Deposit date: | 2022-05-06 | | Release date: | 2023-05-10 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | The structural basis for 2'-5'/3'-5'-cGAMP synthesis by cGAS
Nat Commun, 15, 2024
|
|
