6XSW
 
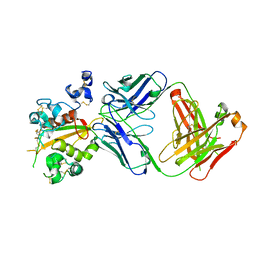 | | Structure of the Notch3 NRR in complex with an antibody Fab Fragment | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Anti-N3 Fab Heavy Chain, ... | | Authors: | Bard, J. | | Deposit date: | 2020-07-16 | | Release date: | 2021-07-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | NOTCH3-targeted antibody drug conjugates regress tumors by inducing apoptosis in receptor cells and through transendocytosis into ligand cells.
Cell Rep Med, 2, 2021
|
|
1FA0
 
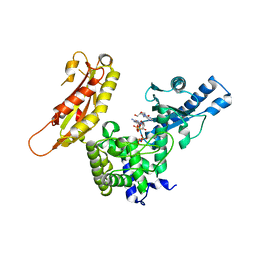 | | STRUCTURE OF YEAST POLY(A) POLYMERASE BOUND TO MANGANATE AND 3'-DATP | | Descriptor: | 3'-DEOXYADENOSINE, 3'-DEOXYADENOSINE-5'-TRIPHOSPHATE, MANGANESE (II) ION, ... | | Authors: | Bard, J, Zhelkovsky, A.M, Helmling, S, Moore, C.L, Bohm, A. | | Deposit date: | 2000-07-11 | | Release date: | 2000-08-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of yeast poly(A) polymerase alone and in complex with 3'-dATP.
Science, 289, 2000
|
|
3LJ3
 
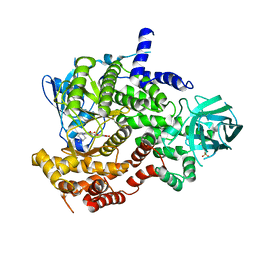 | | PI3-kinase-gamma with a pyrrolopyridine-benzofuran inhibitor | | Descriptor: | (2Z)-4,6-dihydroxy-2-{[1-methyl-4-(4-methylpiperazin-1-yl)-1H-pyrrolo[2,3-b]pyridin-3-yl]methylidene}-1-benzofuran-3(2H)-one, Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma isoform, SULFATE ION | | Authors: | Bard, J, Svenson, K. | | Deposit date: | 2010-01-25 | | Release date: | 2010-04-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Discovery and optimization of 2-(4-substituted-pyrrolo[2,3-b]pyridin-3-yl)methylene-4-hydroxybenzofuran-3(2H)-ones as potent and selective ATP-competitive inhibitors of the mammalian target of rapamycin (mTOR).
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3IBE
 
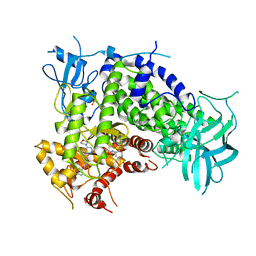 | | Crystal Structure of a Pyrazolopyrimidine Inhibitor Bound to PI3 Kinase Gamma | | Descriptor: | 1-(4-{4-morpholin-4-yl-1-[1-(pyridin-3-ylcarbonyl)piperidin-4-yl]-1H-pyrazolo[3,4-d]pyrimidin-6-yl}phenyl)-3-pyridin-4-ylurea, Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma isoform, SULFATE ION | | Authors: | Bard, J, Svenson, K. | | Deposit date: | 2009-07-15 | | Release date: | 2009-09-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.798 Å) | | Cite: | ATP-Competitive Inhibitors of the Mammalian Target of Rapamycin: Design and Synthesis of Highly Potent and Selective Pyrazolopyrimidines.
J.Med.Chem., 52, 2009
|
|
3H6K
 
 | |
3HFG
 
 | |
3MJW
 
 | | PI3 Kinase gamma with a benzofuranone inhibitor | | Descriptor: | (2Z)-4,6-dihydroxy-2-[(8-methoxy-1,2,3,4-tetrahydropyrazino[1,2-a]indol-10-yl)methylidene]-1-benzofuran-3(2H)-one, Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma isoform | | Authors: | Bard, J, Svenson, K. | | Deposit date: | 2010-04-13 | | Release date: | 2010-06-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | 5-Ureidobenzofuranone indoles as potent and efficacious inhibitors of PI3 kinase-alpha and mTOR for the treatment of breast cancer.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
2Q85
 
 | |
2GC8
 
 | | Structure of a Proline Sulfonamide Inhibitor Bound to HCV NS5b Polymerase | | Descriptor: | 1-[(2-AMINO-4-CHLORO-5-METHYLPHENYL)SULFONYL]-L-PROLINE, RNA-directed RNA polymerase | | Authors: | Gopalsamy, A, Chopra, R, Lim, K, Ciszewski, G, Shi, M, Curran, K.J, Sukits, S.F, Svenson, K, Bard, J, Ellingboe, J.W, Agarwal, A, Krishnamurthy, G, Howe, A.Y, Orlowski, M, Feld, B, O'connell, J, Mansour, T.S. | | Deposit date: | 2006-03-13 | | Release date: | 2006-06-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of Proline Sulfonamides as Potent and Selective Hepatitis C Virus NS5b Polymerase Inhibitors. Evidence for a New NS5b Polymerase Binding Site.
J.Med.Chem., 49, 2006
|
|
5C6W
 
 | | anti-CXCL13 scFv - E10 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Tu, C, Bard, J, Mosyak, L. | | Deposit date: | 2015-06-23 | | Release date: | 2015-11-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Optimization of a scFv-based biotherapeutic by CDR side-chain clash repair
To Be Published
|
|
5CBE
 
 | | E10 in complex with CXCL13 | | Descriptor: | 1,2-ETHANEDIOL, C-X-C motif chemokine 13, E10 heavy chain, ... | | Authors: | Tu, C, Bard, J, Mosyak, L. | | Deposit date: | 2015-06-30 | | Release date: | 2015-11-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A Combination of Structural and Empirical Analyses Delineates the Key Contacts Mediating Stability and Affinity Increases in an Optimized Biotherapeutic Single-chain Fv (scFv).
J. Biol. Chem., 291, 2016
|
|
5CBA
 
 | | 3B4 in complex with CXCL13 - 3B4-CXCL13 | | Descriptor: | 1,2-ETHANEDIOL, 3b4 heavy chain, 3b4 light chain, ... | | Authors: | Tu, C, Bard, J, Mosyak, L. | | Deposit date: | 2015-06-30 | | Release date: | 2015-11-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A Combination of Structural and Empirical Analyses Delineates the Key Contacts Mediating Stability and Affinity Increases in an Optimized Biotherapeutic Single-chain Fv (scFv).
J. Biol. Chem., 291, 2016
|
|
4GLR
 
 | | Structure of the anti-ptau Fab (pT231/pS235_1) in complex with phosphoepitope pT231/pS235 | | Descriptor: | PHOSPHATE ION, anti-ptau heavy chain, anti-ptau light chain, ... | | Authors: | Tu, C, Mosyak, L, Bard, J. | | Deposit date: | 2012-08-14 | | Release date: | 2012-11-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | An Ultra-specific Avian Antibody to Phosphorylated Tau Protein Reveals a Unique Mechanism for Phosphoepitope Recognition.
J.Biol.Chem., 287, 2012
|
|
5C2B
 
 | | anti-CXCL13 parental scFv - 3B4 | | Descriptor: | CHLORIDE ION, scFv 3B4 | | Authors: | Tu, C, Bard, J, Mosyak, L. | | Deposit date: | 2015-06-15 | | Release date: | 2015-11-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.4049 Å) | | Cite: | Optimization of a scFv-based biotherapeutic by CDR side-chain clash repair
To Be Published
|
|
4FUL
 
 | | PI3 Kinase Gamma bound to a pyrmidine inhibitor | | Descriptor: | 4-({4-[3-(piperidin-1-ylcarbonyl)phenyl]pyrimidin-2-yl}amino)benzenesulfonamide, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform | | Authors: | Gopalsamy, A, Bennett, E.M, Shi, M, Zhang, W.G, Bard, J, Yu, K. | | Deposit date: | 2012-06-28 | | Release date: | 2012-10-17 | | Last modified: | 2012-10-31 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Identification of pyrimidine derivatives as hSMG-1 inhibitors.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
1T0R
 
 | | Crystal Structure of the Toluene/o-xylene Monooxygenase Hydroxuylase from Pseudomonas stutzeri-azide bound | | Descriptor: | AZIDE ION, FE (III) ION, HYDROXIDE ION, ... | | Authors: | Sazinsky, M.H, Bard, J, Di Donato, A, Lippard, S.J. | | Deposit date: | 2004-04-12 | | Release date: | 2004-07-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of the Toluene/o-Xylene Monooxygenase Hydroxylase from Pseudomonas stutzeri OX1: INSIGHT INTO THE SUBSTRATE SPECIFICITY, SUBSTRATE CHANNELING, AND ACTIVE SITE TUNING OF MULTICOMPONENT MONOOXYGENASES.
J.Biol.Chem., 279, 2004
|
|
1T0S
 
 | | Structure of the Toluene/o-Xylene Monooxygenase Hydroxylase with 4-bromophenol bound | | Descriptor: | 4-BROMOPHENOL, FE (III) ION, HYDROXIDE ION, ... | | Authors: | Sazinsky, M.H, Bard, J, Di Donato, A, Lippard, S.J. | | Deposit date: | 2004-04-12 | | Release date: | 2004-07-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of the Toluene/o-Xylene Monooxygenase Hydroxylase from Pseudomonas stutzeri OX1: INSIGHT INTO THE SUBSTRATE SPECIFICITY, SUBSTRATE CHANNELING, AND ACTIVE SITE TUNING OF MULTICOMPONENT MONOOXYGENASES.
J.Biol.Chem., 279, 2004
|
|
1T0Q
 
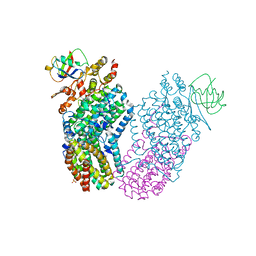 | | Structure of the Toluene/o-Xylene Monooxygenase Hydroxylase | | Descriptor: | FE (III) ION, HYDROXIDE ION, SULFANYLACETIC ACID, ... | | Authors: | Sazinsky, M.H, Bard, J, Di Donato, A, Lippard, S.J. | | Deposit date: | 2004-04-12 | | Release date: | 2004-07-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal Structure of the Toluene/o-Xylene Monooxygenase Hydroxylase from Pseudomonas stutzeri OX1: INSIGHT INTO THE SUBSTRATE SPECIFICITY, SUBSTRATE CHANNELING, AND ACTIVE SITE TUNING OF MULTICOMPONENT MONOOXYGENASES.
J.Biol.Chem., 279, 2004
|
|
1K90
 
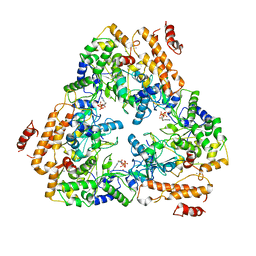 | | Crystal structure of the adenylyl cyclase domain of anthrax edema factor (EF) in complex with calmodulin and 3' deoxy-ATP | | Descriptor: | 3'-DEOXYADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, CALMODULIN, ... | | Authors: | Drum, C.L, Yan, S.-Z, Bard, J, Shen, Y.-Q, Lu, D, Soelaiman, S, Grabarek, Z, Bohm, A, Tang, W.-J. | | Deposit date: | 2001-10-26 | | Release date: | 2002-01-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural basis for the activation of anthrax adenylyl cyclase exotoxin by calmodulin.
Nature, 415, 2002
|
|
1KAQ
 
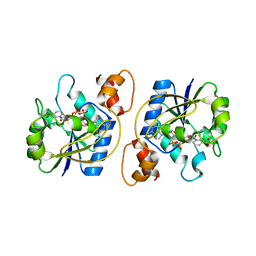 | | Structure of Bacillus subtilis Nicotinic Acid Mononucleotide Adenylyl Transferase | | Descriptor: | NICOTINATE-NUCLEOTIDE ADENYLYLTRANSFERASE, NICOTINIC ACID ADENINE DINUCLEOTIDE | | Authors: | Olland, A.M, Underwood, K.W, Czerwinski, R.M, Lo, M.C, Aulabaugh, A, Bard, J, Stahl, M.L, Somers, W.S, Sullivan, F.X, Chopra, R. | | Deposit date: | 2001-11-02 | | Release date: | 2002-07-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Identification, characterization, and crystal structure of Bacillus subtilis nicotinic acid mononucleotide adenylyltransferase.
J.Biol.Chem., 277, 2002
|
|
1K8T
 
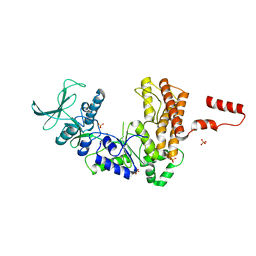 | | Crystal structure of the adenylyl cyclase domain of anthrax edema factor (EF) | | Descriptor: | CALMODULIN-SENSITIVE ADENYLATE CYCLASE, NICKEL (II) ION, SULFATE ION | | Authors: | Drum, C.L, Yan, S.-Z, Bard, J, Shen, Y.-Q, Lu, D, Soelaiman, S, Grabarek, Z, Bohm, A, Tang, W.-J. | | Deposit date: | 2001-10-25 | | Release date: | 2002-01-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for the activation of anthrax adenylyl cyclase exotoxin by calmodulin
Nature, 415, 2002
|
|
1IKO
 
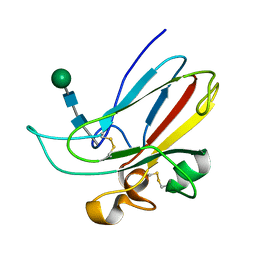 | | CRYSTAL STRUCTURE OF THE MURINE EPHRIN-B2 ECTODOMAIN | | Descriptor: | EPHRIN-B2, alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Toth, J, Cutforth, T, Gelinas, A.D, Bethoney, K.A, Bard, J, Harrison, C.J. | | Deposit date: | 2001-05-03 | | Release date: | 2002-05-03 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Crystal structure of an ephrin ectodomain.
Dev.Cell, 1, 2001
|
|
1K93
 
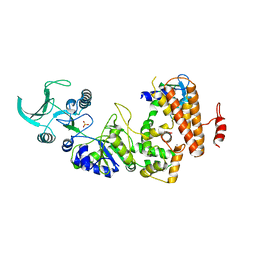 | | Crystal structure of the adenylyl cyclase domain of anthrax edema factor (EF) in complex with calmodulin | | Descriptor: | CALCIUM ION, CALMODULIN, CALMODULIN-SENSITIVE ADENYLATE CYCLASE, ... | | Authors: | Drum, C.L, Yan, S.-Z, Bard, J, Shen, Y.-Q, Lu, D, Soelaiman, S, Grabarek, Z, Bohm, A, Tang, W.-J. | | Deposit date: | 2001-10-26 | | Release date: | 2002-01-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural basis for the activation of anthrax adenylyl cyclase exotoxin by calmodulin.
Nature, 415, 2002
|
|
1KAM
 
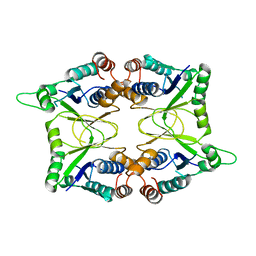 | | Structure of Bacillus subtilis Nicotinic Acid Mononucleotide Adenylyl Transferase | | Descriptor: | NICOTINATE-NUCLEOTIDE ADENYLYLTRANSFERASE | | Authors: | Olland, A.M, Underwood, K.W, Czerwinski, R.M, Lo, M.C, Aulabaugh, A, Bard, J, Stahl, M.L, Somers, W.S, Sullivan, F.X, Chopra, R. | | Deposit date: | 2001-11-02 | | Release date: | 2002-07-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Identification, characterization, and crystal structure of Bacillus subtilis nicotinic acid mononucleotide adenylyltransferase.
J.Biol.Chem., 277, 2002
|
|
2QE5
 
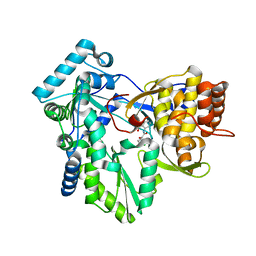 | |
