2AGT
 
 | | Aldose Reductase Mutant Leu 300 Pro complexed with Fidarestat | | Descriptor: | (2S,4S)-2-AMINOFORMYL-6-FLUORO-SPIRO[CHROMAN-4,4'-IMIDAZOLIDINE]-2',5'-DIONE, Aldose reductase, CHLORIDE ION, ... | | Authors: | Petrova, T, Steuber, H, Hazemann, I, Cousido-Siah, A, Mitschler, A, Chung, R, Oka, M, Klebe, G, El-Kabbani, O, Joachimiak, A, Podjarny, A. | | Deposit date: | 2005-07-27 | | Release date: | 2005-09-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Factorizing Selectivity Determinants of Inhibitor Binding toward Aldose and Aldehyde Reductases: Structural and Thermodynamic Properties of the Aldose Reductase Mutant Leu300Pro-Fidarestat Complex
J.Med.Chem., 48, 2005
|
|
6EHN
 
 | | Structural insight into a promiscuous CE15 esterase from the marine bacterial metagenome | | Descriptor: | Carbohydrate esterase MZ0003, GLYCEROL | | Authors: | Helland, R, De Santi, C, Gani, O, Williamson, A.K. | | Deposit date: | 2017-09-13 | | Release date: | 2018-03-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural insight into a CE15 esterase from the marine bacterial metagenome.
Sci Rep, 7, 2017
|
|
9GWT
 
 | | crystal structure of 23ME-00610 Fab in complex with human CD200R1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 23ME-00610 Fab (heavy), 23ME-00610 Fab (light), ... | | Authors: | Huang, Y.M, Ganichkin, O.M. | | Deposit date: | 2024-09-27 | | Release date: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | CD200R1 immune checkpoint blockade by the first-in-human anti-CD200R1 antibody 23ME-00610: molecular mechanism and engineering of a surrogate antibody.
Mabs, 16, 2024
|
|
9GWZ
 
 | | Crystal structure of 23ME-00610 Fab | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 23ME-00610 Fab (heavy), 23ME-00610 Fab (light), ... | | Authors: | Huang, Y.M, Ganichkin, O.M. | | Deposit date: | 2024-09-27 | | Release date: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | CD200R1 immune checkpoint blockade by the first-in-human anti-CD200R1 antibody 23ME-00610: molecular mechanism and engineering of a surrogate antibody.
Mabs, 16, 2024
|
|
3O3R
 
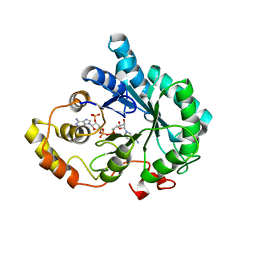 | | Crystal Structure of AKR1B14 in complex with NADP | | Descriptor: | Aldo-keto reductase family 1, member B7, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Sundaram, K, Dhagat, U, El-Kabbani, O. | | Deposit date: | 2010-07-26 | | Release date: | 2011-01-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structure of rat aldose reductase-like protein AKR1B14 holoenzyme: Probing the role of His269 in coenzyme binding by site-directed mutagenesis
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3NTY
 
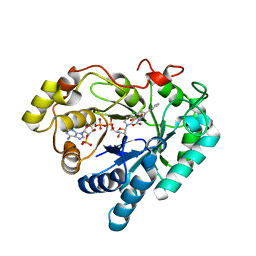 | | Crystal structure of AKR1C1 in complex with NADP and 5-Phenyl,3-chlorosalicylic acid | | Descriptor: | 5-chloro-4-hydroxybiphenyl-3-carboxylic acid, Aldo-keto reductase family 1 member C1, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Dhagat, U, El-Kabbani, O. | | Deposit date: | 2010-07-06 | | Release date: | 2011-04-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Probing the inhibitor selectivity pocket of human 20 alpha-hydroxysteroid dehydrogenase (AKR1C1) with X-ray crystallography and site-directed mutagenesis
Bioorg.Med.Chem.Lett., 21, 2011
|
|
4WKZ
 
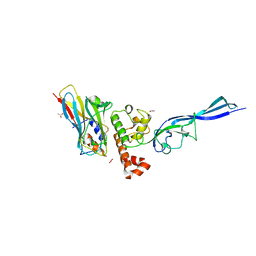 | | Complex of autonomous ScaG cohesin CohG and X-doc domains | | Descriptor: | ACETATE ION, Autonomous cohesin, CALCIUM ION, ... | | Authors: | Voronov-Goldman, M, Lamed, R, Bayer, E.A, Yaniv, O, Shimon, L.J.W. | | Deposit date: | 2014-10-05 | | Release date: | 2015-12-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Complex of autonomous ScaG cohesin CohG and X-doc domains
To Be Published
|
|
3GUG
 
 | |
3H4G
 
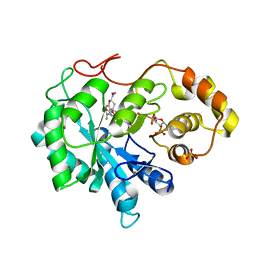 | | Structure of aldehyde reductase holoenzyme in complex with potent aldose reductase inhibitor Fidarestat: Implications for inhibitor binding and selectivity | | Descriptor: | (2S,4S)-2-AMINOFORMYL-6-FLUORO-SPIRO[CHROMAN-4,4'-IMIDAZOLIDINE]-2',5'-DIONE, Alcohol dehydrogenase [NADP+], NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Carbone, V, El-Kabbani, O. | | Deposit date: | 2009-04-20 | | Release date: | 2009-05-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of aldehyde reductase holoenzyme in complex with the potent aldose reductase inhibitor fidarestat: implications for inhibitor binding and selectivity
J.Med.Chem., 48, 2005
|
|
3CV6
 
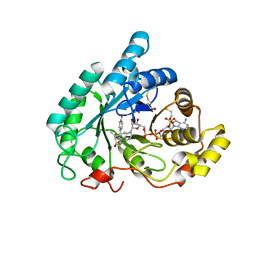 | | The crystal structure of mouse 17-alpha hydroxysteroid dehydrogenase GG225.226PP mutant in complex with inhibitor and cofactor NADP+. | | Descriptor: | 4-[(1R,2S)-1-ethyl-2-(4-hydroxyphenyl)butyl]phenol, Aldo-keto reductase family 1 member C21, BETA-MERCAPTOETHANOL, ... | | Authors: | Dhagat, U, El-Kabbani, O. | | Deposit date: | 2008-04-17 | | Release date: | 2009-03-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the G225P/G226P mutant of mouse 3(17)alpha-hydroxysteroid dehydrogenase (AKR1C21) ternary complex: implications for the binding of inhibitor and substrate.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
3D3W
 
 | |
3CV7
 
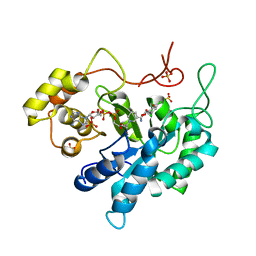 | | Crystal structure of porcine aldehyde reductase ternary complex | | Descriptor: | 3,5-dichloro-2-hydroxybenzoic acid, Alcohol dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Carbone, V, El-Kabbani, O. | | Deposit date: | 2008-04-18 | | Release date: | 2008-10-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.412 Å) | | Cite: | Structure of aldehyde reductase in ternary complex with coenzyme and the potent 20alpha-hydroxysteroid dehydrogenase inhibitor 3,5-dichlorosalicylic acid: Implications for inhibitor binding and selectivity
Arch.Biochem.Biophys., 479, 2008
|
|
3FJN
 
 | |
3FX4
 
 | | Porcine aldehyde reductase in ternary complex with inhibitor | | Descriptor: | Alcohol dehydrogenase [NADP+], NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SULFATE ION, ... | | Authors: | Carbone, V, El-Kabbani, O. | | Deposit date: | 2009-01-20 | | Release date: | 2009-12-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structure of aldehyde reductase in ternary complex with a 5-arylidene-2,4-thiazolidinedione aldose reductase inhibitor.
Eur.J.Med.Chem., 45, 2010
|
|
1WNT
 
 | | Structure of the tetrameric form of Human L-Xylulose Reductase | | Descriptor: | L-xylulose reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | El-Kabbani, O, Carbone, V, Darmanin, C, Ishikura, S, Hara, A. | | Deposit date: | 2004-08-09 | | Release date: | 2005-07-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the tetrameric form of human L-Xylulose reductase: Probing the inhibitor-binding site with molecular modeling and site-directed mutagenesis
Proteins, 60, 2005
|
|
3OHS
 
 | |
1X98
 
 | | Crystal structure of Aldose Reductase complexed with 2S4R (Stereoisomer of Fidarestat, 2S4S) | | Descriptor: | (2S,4R)-2-AMINOFORMYL-6-FLUORO-SPIRO[CHROMAN-4,4'-IMIDAZOLIDINE]-2',5'-DIONE, Aldose Reductase, CITRIC ACID, ... | | Authors: | El-Kabbani, O, Darmanin, C, Oka, M, Schulze-Briese, C, Tomizaki, T, Hazemann, I, Mitschler, A, Podjarny, A. | | Deposit date: | 2004-08-19 | | Release date: | 2004-09-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | High-Resolution Structures of Human Aldose Reductase Holoenzyme in Complex with Stereoisomers of the Potent Inhibitor Fidarestat: Stereospecific Interaction between the Enzyme and a Cyclic Imide Type Inhibitor
J.Med.Chem., 47, 2004
|
|
3QKZ
 
 | | Crystal structure of mutant His269Arg AKR1B14 | | Descriptor: | Aldo-keto reductase family 1, member B7, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Sundaram, K, El-Kabbani, O. | | Deposit date: | 2011-02-02 | | Release date: | 2012-02-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structure of the His269Arg mutant of the rat aldose reductase-like protein AKR1B14 complexed with NADPH.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
1X97
 
 | | Crystal structure of Aldose Reductase complexed with 2R4S (Stereoisomer of Fidarestat, 2S4S) | | Descriptor: | (2R,4S)-2-AMINOFORMYL-6-FLUORO-SPIRO[CHROMAN-4,4'-IMIDAZOLIDINE]-2',5'-DIONE, Aldose Reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | El-Kabbani, O, Darmanin, C, Oka, M, Schulze-Briese, C, Tomizaki, T, Hazemann, I, Mitschler, A, Podjarny, A. | | Deposit date: | 2004-08-19 | | Release date: | 2004-09-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | High-Resolution Structures of Human Aldose Reductase Holoenzyme in Complex with Stereoisomers of the Potent Inhibitor Fidarestat: Stereospecific Interaction between the Enzyme and a Cyclic Imide Type Inhibitor
J.Med.Chem., 47, 2004
|
|
1X96
 
 | | Crystal structure of Aldose Reductase with citrates bound in the active site | | Descriptor: | CITRIC ACID, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, aldose reductase | | Authors: | El-Kabbani, O, Darmanin, C, Oka, M, Schulze-Briese, C, Tomizaki, T, Hazemann, I, Mitschler, A, Podjarny, A. | | Deposit date: | 2004-08-19 | | Release date: | 2004-09-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | High-Resolution Structures of Human Aldose Reductase Holoenzyme in Complex with Stereoisomers of the Potent Inhibitor Fidarestat: Stereospecific Interaction between the Enzyme and a Cyclic Imide Type Inhibitor
J.Med.Chem., 47, 2004
|
|
8AH3
 
 | |
2ALR
 
 | | ALDEHYDE REDUCTASE | | Descriptor: | ALDEHYDE REDUCTASE | | Authors: | El-Kabbani, O. | | Deposit date: | 1994-09-06 | | Release date: | 1996-06-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Structures of human and porcine aldehyde reductase: an enzyme implicated in diabetic complications.
Acta Crystallogr.,Sect.D, 50, 1994
|
|
1AE4
 
 | |
1C8T
 
 | | HUMAN STROMELYSIN-1 (E202Q) CATALYTIC DOMAIN COMPLEXED WITH RO-26-2812 | | Descriptor: | 2-(2-{2-[(BIPHENYL-4-YLMETHYL)-AMINO]-3-MERCAPTO-PENTANOYLAMINO}-ACETYLAMINO)-3-METHYL-BUTYRIC ACID METHYL ESTER, CALCIUM ION, STROMELYSIN-1, ... | | Authors: | Steele, D.L, el-Kabbani, O, Dunten, P, Crowther, R.L. | | Deposit date: | 1999-07-29 | | Release date: | 2000-07-19 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Expression, characterization and structure determination of an active site mutant (Glu202-Gln) of mini-stromelysin-1.
Protein Eng., 13, 2000
|
|
3MDF
 
 | | Crystal structure of the RRM domain of Cyclophilin 33 | | Descriptor: | Peptidyl-prolyl cis-trans isomerase E | | Authors: | Hom, R.A, Chang, P.Y, Roy, S, Mussleman, C.A, Glass, K.C, Seleznevia, A.I, Gozani, O, Ismagilov, R.F, Cleary, M.L, Kutateladze, T.G. | | Deposit date: | 2010-03-30 | | Release date: | 2010-05-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Molecular mechanism of MLL PHD3 and RNA recognition by the Cyp33 RRM domain.
J.Mol.Biol., 400, 2010
|
|
