8K1F
 
 | |
8K1C
 
 | |
8K76
 
 | |
4NPQ
 
 | | The resting-state conformation of the GLIC ligand-gated ion channel | | Descriptor: | Proton-gated ion channel | | Authors: | Sauguet, L, Shahsavar, A, Poitevin, F, Huon, C, Menny, A, Nemecz, A, Haouz, A, Changeux, J.P, Corringer, P.J, Delarue, M. | | Deposit date: | 2013-11-22 | | Release date: | 2013-12-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (4.35 Å) | | Cite: | Crystal structures of a pentameric ligand-gated ion channel provide a mechanism for activation.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
5T09
 
 | | The structure of the type III effector HopBA1 | | Descriptor: | CHLORIDE ION, POTASSIUM ION, Type III secretion system effector HopBA1 | | Authors: | Cherkis, K, Machius, M, Nishimura, M.T, Dangl, J.L. | | Deposit date: | 2016-08-15 | | Release date: | 2017-02-01 | | Last modified: | 2019-11-20 | | Method: | X-RAY DIFFRACTION (2.013 Å) | | Cite: | TIR-only protein RBA1 recognizes a pathogen effector to regulate cell death in Arabidopsis.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
7EKZ
 
 | | Structural and functional insights into Hydra Actinoporin-Like Toxin 1 (HALT-1) | | Descriptor: | FORMIC ACID, GLYCEROL, HALT-1 | | Authors: | Ker, D.S, Sha, X.H, Jonet, M.A, Hwang, J.S, Ng, C.L. | | Deposit date: | 2021-04-07 | | Release date: | 2022-02-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Structural and functional analysis of Hydra Actinoporin-Like Toxin 1 (HALT-1).
Sci Rep, 11, 2021
|
|
3BK3
 
 | |
1HF9
 
 | | C-Terminal Coiled-Coil Domain from Bovine IF1 | | Descriptor: | ATPASE INHIBITOR (MITOCHONDRIAL) | | Authors: | Gordon-Smith, D.J, Carbajo, R.J, Yang, J.-C, Videler, H, Runswick, M.J, Walker, J.E, Neuhaus, D. | | Deposit date: | 2000-11-30 | | Release date: | 2001-05-31 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a C-terminal coiled-coil domain from bovine IF(1): the inhibitor protein of F(1) ATPase.
J. Mol. Biol., 308, 2001
|
|
4MX7
 
 | | Structure of mouse CD1d in complex with dioleoyl-phosphatidic acid | | Descriptor: | (2R)-3-(phosphonooxy)propane-1,2-diyl (9Z,9'Z)bis-octadec-9-enoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zajonc, D.M, Wang, J. | | Deposit date: | 2013-09-25 | | Release date: | 2014-10-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | TBN
To be Published
|
|
2PPH
 
 | | solution structure of human MEKK3 PB1 domain | | Descriptor: | Mitogen-activated protein kinase kinase kinase 3 | | Authors: | Hu, Q, Zhang, J, Wu, J, Shi, Y. | | Deposit date: | 2007-04-30 | | Release date: | 2007-05-22 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Insight into the Binding Properties of MEKK3 PB1 to MEK5 PB1 from Its Solution Structure.
Biochemistry, 46, 2007
|
|
3Q2M
 
 | | Crystal Structure of Spectinomycin Phosphotransferase, APH(9)-Ia, Protein Kinase Inhibitor CKI-7 Complex | | Descriptor: | N-(2-AMINOETHYL)-5-CHLOROISOQUINOLINE-8-SULFONAMIDE, NICKEL (II) ION, Spectinomycin phosphotransferase | | Authors: | Berghuis, A.M, Fong, D.H, Xiong, B, Hwang, J. | | Deposit date: | 2010-12-20 | | Release date: | 2011-05-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structures of two aminoglycoside kinases bound with a eukaryotic protein kinase inhibitor.
Plos One, 6, 2011
|
|
1KUY
 
 | | X-ray Crystallographic Studies of Serotonin N-acetyltransferase Catalysis and Inhibition | | Descriptor: | COA-S-ACETYL TRYPTAMINE, Serotonin N-acetyltransferase | | Authors: | Wolf, E, De Angelis, J, Khalil, E.M, Cole, P.A, Burley, S.K. | | Deposit date: | 2002-01-22 | | Release date: | 2002-03-22 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | X-ray crystallographic studies of serotonin N-acetyltransferase catalysis and inhibition.
J.Mol.Biol., 317, 2002
|
|
1I5T
 
 | | SOLUTION STRUCTURE OF CYANOFERRICYTOCHROME C | | Descriptor: | CYANIDE ION, CYTOCHROME C, HEME C | | Authors: | Yao, Y, Qian, C, Ye, K, Wang, J, Tang, W. | | Deposit date: | 2001-02-28 | | Release date: | 2001-03-21 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of cyanoferricytochrome c: ligand-controlled conformational flexibility and electronic structure of the heme moiety.
J.Biol.Inorg.Chem., 7, 2002
|
|
5OF4
 
 | | The cryo-EM structure of human TFIIH | | Descriptor: | General transcription factor IIH subunit 2, General transcription factor IIH subunit 3, General transcription factor IIH subunit 4,p52,General transcription factor IIH subunit 4, ... | | Authors: | Greber, B.J, Nguyen, T.H.D, Fang, J, Afonine, P.V, Adams, P.D, Nogales, E. | | Deposit date: | 2017-07-10 | | Release date: | 2017-09-13 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | The cryo-electron microscopy structure of human transcription factor IIH.
Nature, 549, 2017
|
|
3S2A
 
 | | Crystal structure of PI3K-gamma in complex with a quinoline inhibitor | | Descriptor: | N-{2-chloro-5-[4-(morpholin-4-yl)quinolin-6-yl]pyridin-3-yl}-4-fluorobenzenesulfonamide, Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma isoform, SULFATE ION | | Authors: | Whittington, D.A, Tang, J, Yakowec, P. | | Deposit date: | 2011-05-16 | | Release date: | 2011-06-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Phospshoinositide 3-Kinase (PI3K)/Mammalian Target of Rapamycin (mTOR) Dual Inhibitors: Discovery and Structure-Activity Relationships of a Series of Quinoline and Quinoxaline Derivatives.
J.Med.Chem., 54, 2011
|
|
2Q7N
 
 | | Crystal structure of Leukemia inhibitory factor in complex with LIF receptor (domains 1-5) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Huyton, T, Zhang, J.G, Nicola, N.A, Garrett, T.P.J. | | Deposit date: | 2007-06-07 | | Release date: | 2007-07-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | An unusual cytokine:Ig-domain interaction revealed in the crystal structure of leukemia inhibitory factor (LIF) in complex with the LIF receptor.
Proc.Natl.Acad.Sci.USA, 104, 2007
|
|
4NPP
 
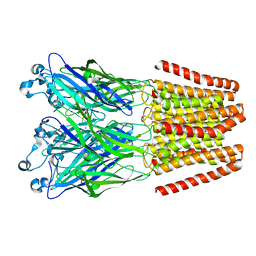 | | The GLIC-His10 wild-type structure in equilibrium between the open and locally-closed (LC) forms | | Descriptor: | NICKEL (II) ION, Proton-gated ion channel | | Authors: | Sauguet, L, Shahsavar, A, Poitevin, F, Huon, C, Menny, A, Nemecz, A, Haouz, A, Changeux, J.P, Corringer, P.J, Delarue, M. | | Deposit date: | 2013-11-22 | | Release date: | 2013-12-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Crystal structures of a pentameric ligand-gated ion channel provide a mechanism for activation.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
2FXZ
 
 | |
7KJX
 
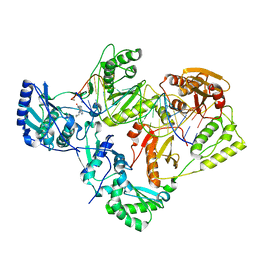 | | Structure of HIV-1 reverse transcriptase initiation complex core with nevirapine | | Descriptor: | 11-CYCLOPROPYL-5,11-DIHYDRO-4-METHYL-6H-DIPYRIDO[3,2-B:2',3'-E][1,4]DIAZEPIN-6-ONE, HIV-1 viral RNA fragment, MAGNESIUM ION, ... | | Authors: | Ha, B, Larsen, K.P, Zhang, J, Fu, Z, Montabana, E, Jackson, L.N, Chen, D.H, Puglisi, E.V. | | Deposit date: | 2020-10-26 | | Release date: | 2021-03-17 | | Last modified: | 2021-05-12 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | High-resolution view of HIV-1 reverse transcriptase initiation complexes and inhibition by NNRTI drugs.
Nat Commun, 12, 2021
|
|
7KJW
 
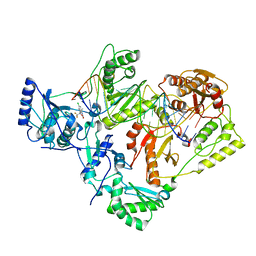 | | Structure of HIV-1 reverse transcriptase initiation complex core with efavirenz | | Descriptor: | (-)-6-CHLORO-4-CYCLOPROPYLETHYNYL-4-TRIFLUOROMETHYL-1,4-DIHYDRO-2H-3,1-BENZOXAZIN-2-ONE, HIV-1 viral RNA fragment, MAGNESIUM ION, ... | | Authors: | Ha, B, Larsen, K.P, Zhang, J, Fu, Z, Montabana, E, Jackson, L.N, Chen, D.H, Puglisi, E.V. | | Deposit date: | 2020-10-26 | | Release date: | 2021-03-17 | | Last modified: | 2021-05-12 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | High-resolution view of HIV-1 reverse transcriptase initiation complexes and inhibition by NNRTI drugs.
Nat Commun, 12, 2021
|
|
7KJV
 
 | | Structure of HIV-1 reverse transcriptase initiation complex core | | Descriptor: | HIV-1 viral RNA fragment, MAGNESIUM ION, Reverse transcriptase/ribonuclease H, ... | | Authors: | Ha, B, Larsen, K.P, Zhang, J, Fu, Z, Montabana, E, Jackson, L.N, Chen, D.H, Puglisi, E.V. | | Deposit date: | 2020-10-26 | | Release date: | 2021-03-17 | | Last modified: | 2021-05-12 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | High-resolution view of HIV-1 reverse transcriptase initiation complexes and inhibition by NNRTI drugs.
Nat Commun, 12, 2021
|
|
2GF9
 
 | | Crystal structure of human RAB3D in complex with GDP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Ras-related protein Rab-3D, ... | | Authors: | Hong, B, Wang, J, Shen, L, Tempel, W, Landry, R, Arrowsmith, C.H, Edwards, A.M, Sundstrom, M, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-03-21 | | Release date: | 2006-05-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Crystal structure of human RAB3D in complex with GDP
To be Published
|
|
1V9P
 
 | | Crystal Structure Of Nad+-Dependent DNA Ligase | | Descriptor: | ADENOSINE MONOPHOSPHATE, DNA ligase, ZINC ION | | Authors: | Lee, J.Y, Chang, C, Song, H.K, Moon, J, Yang, J.K, Kim, H.K, Kwon, S.K, Suh, S.W. | | Deposit date: | 2004-01-27 | | Release date: | 2004-03-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of NAD(+)-dependent DNA ligase: modular architecture and functional implications.
Embo J., 19, 2000
|
|
1UT9
 
 | | Structural Basis for the Exocellulase Activity of the Cellobiohydrolase CbhA from C. thermocellum | | Descriptor: | CELLULOSE 1,4-BETA-CELLOBIOSIDASE | | Authors: | Schubot, F.D, Kataeva, I.A, Chang, J, Shah, A.K, Ljungdahl, L.G, Rose, J.P, Wang, B.C. | | Deposit date: | 2003-12-04 | | Release date: | 2004-02-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for the exocellulase activity of the cellobiohydrolase CbhA from Clostridium thermocellum.
Biochemistry, 43, 2004
|
|
2ZKJ
 
 | | Crystal structure of human PDK4-ADP complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Kato, M, Wynn, R.M, Chuang, J.L, Tso, S.-C, Li, J, Chuang, D.T. | | Deposit date: | 2008-03-25 | | Release date: | 2008-08-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Pyruvate Dehydrogenase Kinase-4 Structures Reveal a Metastable Open Conformation Fostering Robust Core-free Basal Activity
J.Biol.Chem., 283, 2008
|
|
