5FLZ
 
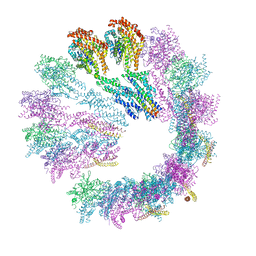 | | Cryo-EM structure of gamma-TuSC oligomers in a closed conformation | | Descriptor: | SPINDLE POLE BODY COMPONENT 110, SPINDLE POLE BODY COMPONENT SPC97, SPINDLE POLE BODY COMPONENT SPC98, ... | | Authors: | Greenberg, C.H, Kollman, J, Zelter, A, Johnson, R, MacCoss, M.J, Davis, T.N, Agard, D.A, Sali, A. | | Deposit date: | 2015-10-29 | | Release date: | 2016-01-13 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (6.9 Å) | | Cite: | Structure of Gamma-Tubulin Small Complex Based on a Cryo-Em Map, Chemical Cross-Links, and a Remotely Related Structure.
J.Struct.Biol., 194, 2016
|
|
7KES
 
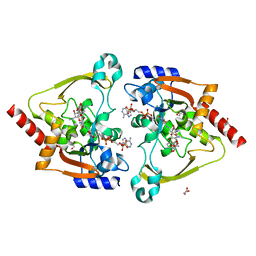 | | Crystal structure of meta-AAC0038, an environmental aminoglycoside resistance enzyme, mutant H168A in complex with apramycin and CoA | | Descriptor: | APRAMYCIN, Aminoglycoside N(3)-acetyltransferase, CHLORIDE ION, ... | | Authors: | Stogios, P.J, Skarina, T, Michalska, K, Xu, Z, Yim, V, Savchenko, A, Joachimiak, A, Satchell, K.J, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-10-12 | | Release date: | 2020-10-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Structural and molecular rationale for the diversification of resistance mediated by the Antibiotic_NAT family.
Commun Biol, 5, 2022
|
|
7EAY
 
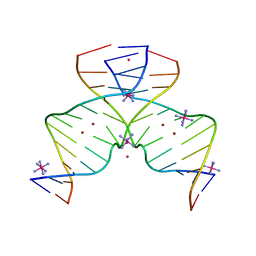 | | DNA containing Cu(II)-mediated 4-N-carboxymethylcytosine base pairs | | Descriptor: | COBALT (II) ION, COBALT HEXAMMINE(III), COPPER (II) ION, ... | | Authors: | Kondo, J, Terashima, A, Yoshimura, A, Tada, Y, Ono, A. | | Deposit date: | 2021-03-08 | | Release date: | 2022-03-09 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | DNA containing Cu(II)-mediated 4-N-carboxymethylcytosine base pairs
To Be Published
|
|
7KEH
 
 | | Crystal structure from SARS-CoV-2 NendoU NSP15 | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, SULFATE ION, Uridylate-specific endoribonuclease | | Authors: | Godoy, A.S, Nakamura, A.M, Pereira, H.M, Noske, G.D, Gawriljuk, V.O, Fernandes, R.S, Oliveira, K.I.Z, Oliva, G. | | Deposit date: | 2020-10-10 | | Release date: | 2020-12-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Allosteric regulation and crystallographic fragment screening of SARS-CoV-2 NSP15 endoribonuclease.
Nucleic Acids Res., 2023
|
|
5FYU
 
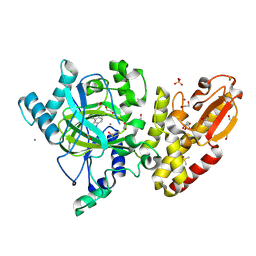 | | Crystal structure of the catalytic domain of human JARID1B in complex with 3D fragment 3-Amino-4-methyl-1,3-dihydro-2H-indol-2-one (N10042a) | | Descriptor: | (3R)-3-azanyl-4-methyl-1,3-dihydroindol-2-one, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Nowak, R, Krojer, T, Johansson, C, Pearce, N, Gileadi, C, Kupinska, K, Strain-Damerell, C, Szykowska, A, Burgess-Brown, N.A, Arrowsmith, C.H, Bountra, C, Edwards, A.M, von Delft, F, Brennan, P.E, Oppermann, U. | | Deposit date: | 2016-03-09 | | Release date: | 2016-03-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Crystal Structure of the Catalytic Domain of Human Jarid1B in Complex with N10042A
To be Published
|
|
5FZM
 
 | | Crystal structure of the catalytic domain of human JARID1B in complex with 3D fragment 5-(2-fluorophenyl)-1,3-oxazole-4-carboxylic acid (N09989b) (ligand modelled based on PANDDA event map, SGC - Diamond I04-1 fragment screening) | | Descriptor: | 1,2-ETHANEDIOL, 5-(2-fluorophenyl)-1,3-oxazole-4-carboxylic acid, CHLORIDE ION, ... | | Authors: | Nowak, R, Krojer, T, Johansson, C, Kupinska, K, Szykowska, A, Pearce, N, Talon, R, Collins, P, Gileadi, C, Strain-Damerell, C, Burgess-Brown, N.A, Arrowsmith, C.H, Bountra, C, Edwards, A.M, von Delft, F, Brennan, P.E, Oppermann, U. | | Deposit date: | 2016-03-14 | | Release date: | 2017-03-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Crystal Structure of the Catalytic Domain of Human Jarid1B in Complex with 3D Fragment 5-(2-Fluorophenyl)-1,3-Oxazole-4-Carboxylic Acid (N09989B) (Ligand Modelled Based on Pandda Event Map, Sgc - Diamond I04-1 Fragment Screening)
To be Published
|
|
5FQJ
 
 | | W229D mutant of the last common ancestor of Gram-negative bacteria (GNCA) beta-lactamase bound to 5(6)-nitrobenzotriazole (TS-analog) | | Descriptor: | 6-NITROBENZOTRIAZOLE, GNCA LACTAMASE W229D | | Authors: | Gavira, J.A, Martinez-Rodriguez, S, Risso, V.A, Sanchez-Ruiz, J.M. | | Deposit date: | 2015-12-11 | | Release date: | 2016-12-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.271 Å) | | Cite: | De novo active sites for resurrected Precambrian enzymes.
Nat Commun, 8, 2017
|
|
5FZ8
 
 | | Crystal structure of the catalytic domain of human JARID1B in complex with malate | | Descriptor: | (2S)-2-hydroxybutanedioic acid, 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Nowak, R, Krojer, T, Johansson, C, Gileadi, C, Kupinska, K, Strain-Damerell, C, Szykowska, A, von Delft, F, Burgess-Brown, N.A, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Oppermann, U. | | Deposit date: | 2016-03-11 | | Release date: | 2017-03-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Crystal Structure of the Catalytic Domain of Human Jarid1B in Complex with Malate
To be Published
|
|
5T5H
 
 | | Structure and assembly model for the Trypanosoma cruzi 60S ribosomal subunit | | Descriptor: | 40S ribosomal protein L14, 5.8S rRNA, 5S rRNA, ... | | Authors: | Liu, Z, Gutierrez-Vargas, C, Wei, J, Grassucci, R.A, Ramesh, M, Espina, N, Sun, M, Tutuncuoglu, B, Madison-Antenucci, S, Woolford Jr, J.L, Tong, L, Frank, J. | | Deposit date: | 2016-08-31 | | Release date: | 2016-10-12 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (2.54 Å) | | Cite: | Structure and assembly model for the Trypanosoma cruzi 60S ribosomal subunit.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5FVN
 
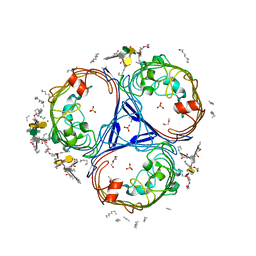 | | X-ray crystal structure of Enterobacter cloacae OmpE36 porin. | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, 3-HYDROXY-TETRADECANOIC ACID, 3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid, ... | | Authors: | Arunmanee, W, Pathania, M, Soloyova, A, Brun, A, Ridley, H, Basle, A, van den Berg, B, Lakey, J.H. | | Deposit date: | 2016-02-09 | | Release date: | 2016-08-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Gram-negative trimeric porins have specific LPS binding sites that are essential for porin biogenesis.
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
5F8K
 
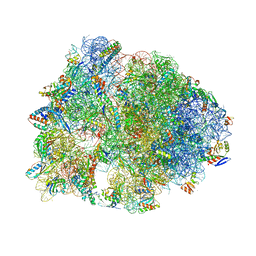 | | Crystal structure of the Bac7(1-16) antimicrobial peptide bound to the Thermus thermophilus 70S ribosome | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Seefeldt, A.C, Graf, M, Perebaskine, N, Nguyen, F, Arenz, S, Mardirossian, M, Scocchi, M, Wilson, D.N, Innis, C.A. | | Deposit date: | 2015-12-09 | | Release date: | 2016-02-03 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the mammalian antimicrobial peptide Bac7(1-16) bound within the exit tunnel of a bacterial ribosome.
Nucleic Acids Res., 44, 2016
|
|
4NXN
 
 | | Crystal Structure of the 30S ribosomal subunit from a GidB (RsmG) mutant of Thermus thermophilus (HB8), bound with streptomycin | | Descriptor: | 16S rRNA, MAGNESIUM ION, STREPTOMYCIN, ... | | Authors: | Demirci, H, Murphy IV, F, Murphy, E, Gregory, S.T, Dahlberg, A.E, Jogl, G. | | Deposit date: | 2013-12-09 | | Release date: | 2014-05-21 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (3.544 Å) | | Cite: | A structural basis for streptomycin resistance
To be Published
|
|
5FQP
 
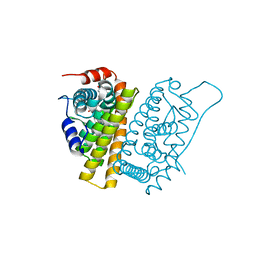 | | Selective estrogen receptor downregulator antagonists: Tetrahydroisoquinoline phenols 1. | | Descriptor: | (E)-3-[4-[(1R,3R)-6-hydroxy-2-isobutyl-3-methyl-3,4-dihydro-1H-isoquinolin-1-yl]phenyl]prop-2-enoic acid, ESTROGEN RECEPTOR ALPHA | | Authors: | Scott, J.S, Bailey, A, Davies, R.D.M, Degorce, S.L, MacFaul, P.A, Gingell, H, Moss, T, Norman, R.A, Pink, J.H, Rabow, A.A, Roberts, B, Smith, P.D. | | Deposit date: | 2015-12-14 | | Release date: | 2016-02-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Tetrahydroisoquinoline Phenols: Selective Estrogen Receptor Downregulator Antagonists with Oral Bioavailability in Rat.
Acs Med.Chem.Lett., 7, 2016
|
|
5FZ0
 
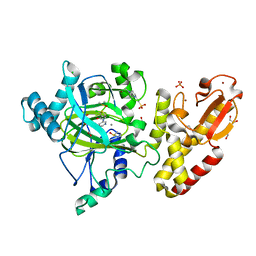 | | Crystal structure of the catalytic domain of human JARID1B in complex with 2,5-dichloro-N-(pyridin-3-yl)thiophene-3-carboxamide (N08137b) (ligand modelled based on PANDDA event map, SGC - Diamond I04-1 fragment screening) | | Descriptor: | 1,2-ETHANEDIOL, 2,5-dichloro-N-(pyridin-3-yl)thiophene-3-carboxamide, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Nowak, R, Krojer, T, Pearce, N, Johansson, C, Gileadi, C, Kupinska, K, Strain-Damerell, C, Szykowska, A, Burgess-Brown, N.A, Arrowsmith, C.H, Bountra, C, Edwards, A.M, von Delft, F, Brennan, P.E, Oppermann, U. | | Deposit date: | 2016-03-10 | | Release date: | 2016-04-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Crystal Structure of the Catalytic Domain of Human Jarid1B in Complex with N11213A
To be Published
|
|
1K9M
 
 | | Co-crystal structure of tylosin bound to the 50S ribosomal subunit of Haloarcula marismortui | | Descriptor: | 23S RRNA, 5S RRNA, CADMIUM ION, ... | | Authors: | Hansen, J.L, Ippolito, J.A, Ban, N, Nissen, P, Moore, P.B, Steitz, T.A. | | Deposit date: | 2001-10-29 | | Release date: | 2002-07-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The structures of four macrolide antibiotics bound to the large ribosomal subunit.
Mol.Cell, 10, 2002
|
|
6MN5
 
 | | Crystal structure of aminoglycoside acetyltransferase AAC(3)-IVa, H154A mutant, in complex with gentamicin C1A | | Descriptor: | (2R,3R,4R,5R)-2-((1S,2S,3R,4S,6R)-4,6-DIAMINO-3-((2R,3R,6S)-3-AMINO-6-(AMINOMETHYL)-TETRAHYDRO-2H-PYRAN-2-YLOXY)-2-HYDR OXYCYCLOHEXYLOXY)-5-METHYL-4-(METHYLAMINO)-TETRAHYDRO-2H-PYRAN-3,5-DIOL, 1,2-ETHANEDIOL, 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, ... | | Authors: | Stogios, P.J, Evdokimova, E, Kim, Y, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-10-01 | | Release date: | 2018-10-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Structural and molecular rationale for the diversification of resistance mediated by the Antibiotic_NAT family.
Commun Biol, 5, 2022
|
|
5FZ9
 
 | | Crystal structure of the catalytic domain of human JARID1B in complex with Maybridge fragment thieno(3,2-b)thiophene-5-carboxylic acid (N06263b) (ligand modelled based on PANDDA event map, SGC - Diamond I04-1 fragment screening) | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Nowak, R, Krojer, T, Johansson, C, Kupinska, K, Szykowska, A, Pearce, N, Talon, R, Collins, P, Gileadi, C, Strain-Damerell, C, Burgess-Brown, N.A, Arrowsmith, C.H, Bountra, C, Edwards, A.M, von Delft, F, Brennan, P.E, Oppermann, U. | | Deposit date: | 2016-03-12 | | Release date: | 2016-03-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Crystal Structure of the Catalytic Domain of Human Jarid1B in Complex with Maybridge Fragment Thieno(3,2-B)Thiophene -5-Carboxylic Acid (N06263B) (Ligand Modelled Based on Pandda Event Map, Sgc - Diamond I04-1 Fragment Screening)
To be Published
|
|
5FZH
 
 | | Crystal structure of the catalytic domain of human JARID1B in complex with Maybridge fragment 4,5-dihydronaphtho(1,2-b)thiophene-2- carboxylicacid (N11181a) (ligand modelled based on PANDDA event map, SGC - Diamond I04-1 fragment screening) | | Descriptor: | (3R)-5-fluoro-3-hydroxy-1,3-dihydro-2H-indol-2-one, 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Nowak, R, Krojer, T, Johansson, C, Kupinska, K, Szykowska, A, Pearce, N, Talon, R, Collins, P, Gileadi, C, Strain-Damerell, C, Burgess-Brown, N.A, Arrowsmith, C.H, Bountra, C, Edwards, A.M, von Delft, F, Brennan, P.E, Oppermann, U. | | Deposit date: | 2016-03-14 | | Release date: | 2016-05-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Crystal Structure of the Catalytic Domain of Human Jarid1B in Complex with Maybridge Fragment 4,5-Dihydronaphtho(1,2-B)Thiophene-2-Carboxylicacid (N11181A) (Ligand Modelled Based on Pandda Event Map, Sgc - Diamond I04-1 Fragment Screening)
To be Published
|
|
3K3Q
 
 | | Crystal Structure of a Llama Antibody complexed with the C. Botulinum Neurotoxin Serotype A Catalytic Domain | | Descriptor: | Botulinum neurotoxin type A, ZINC ION, llama Aa1 VHH domain | | Authors: | Thompson, A.A, Dong, J, Marks, J.D, Stevens, R.C. | | Deposit date: | 2009-10-04 | | Release date: | 2010-02-23 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A Single-Domain Llama Antibody Potently Inhibits the Enzymatic Activity of Botulinum Neurotoxin by Binding to the Non-Catalytic alpha-Exosite Binding Region.
J.Mol.Biol., 397, 2010
|
|
6MTP
 
 | | Crystal structure of VRC42.04 Fab in complex with gp41 peptide | | Descriptor: | Antibody VRC42.04 Fab heavy chain, Antibody VRC42.04 Fab light chain, RV217 founder virus gp41 peptide | | Authors: | Kwon, Y.D, Druz, A, Law, W.H, Peng, D, Zhang, B, Doria-Rose, N.A, Kwong, P.D. | | Deposit date: | 2018-10-21 | | Release date: | 2019-03-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.036 Å) | | Cite: | Longitudinal Analysis Reveals Early Development of Three MPER-Directed Neutralizing Antibody Lineages from an HIV-1-Infected Individual.
Immunity, 50, 2019
|
|
3K9F
 
 | | Detailed structural insight into the quinolone-DNA cleavage complex of type IIA topoisomerases | | Descriptor: | (3S)-9-fluoro-3-methyl-10-(4-methylpiperazin-1-yl)-7-oxo-2,3-dihydro-7H-[1,4]oxazino[2,3,4-ij]quinoline-6-carboxylic acid, DNA (5'-D(*AP*CP*CP*AP*AP*GP*GP*T*CP*AP*TP*GP*AP*AP*T)-3'), DNA (5'-D(*CP*TP*GP*TP*TP*TP*TP*A*CP*GP*TP*GP*CP*AP*T)-3'), ... | | Authors: | Laponogov, I, Pan, X.-S, Veselkov, D.A, Fisher, L.M, Sanderson, M.R. | | Deposit date: | 2009-10-15 | | Release date: | 2009-10-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural Basis of Gate-DNA Breakage and Resealing by Type II Topoisomerases
Plos One, 5, 2010
|
|
4OX9
 
 | | Crystal structure of the aminoglycoside resistance methyltransferase NpmA bound to the 30S ribosomal subunit | | Descriptor: | 16S rRNA, 16S rRNA (adenine(1408)-N(1))-methyltransferase, 30S ribosomal protein S10, ... | | Authors: | Dunkle, J.A, Conn, G.L, Dunham, C.M. | | Deposit date: | 2014-02-04 | | Release date: | 2014-04-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.8035 Å) | | Cite: | Molecular recognition and modification of the 30S ribosome by the aminoglycoside-resistance methyltransferase NpmA.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
5T7V
 
 | | Methicillin Resistant, Linezolid resistant Staphylococcus aureus 70S ribosome (delta S145 uL3) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Belousoff, M.J, Lithgow, T, Eyal, Z, Yonath, A, Radjainia, M. | | Deposit date: | 2016-09-06 | | Release date: | 2017-05-31 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural Basis for Linezolid Binding Site Rearrangement in theStaphylococcus aureusRibosome.
MBio, 8, 2017
|
|
5TBW
 
 | | Crystal structure of chlorolissoclimide bound to the yeast 80S ribosome | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0-A, ... | | Authors: | Konst, Z.A, Szklarski, A.R, Pellegrino, S, Michalak, S.E, Meyer, M, Zanette, C, Cencic, R, Nam, S, Horne, D.A, Pelletier, J, Mobley, D.L, Yusupova, G, Yusupov, M, Vanderwal, C.D. | | Deposit date: | 2016-09-13 | | Release date: | 2017-07-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Synthesis facilitates an understanding of the structural basis for translation inhibition by the lissoclimides.
Nat Chem, 9, 2017
|
|
3FWO
 
 | | The large ribosomal subunit from Deinococcus radiodurans complexed with Methymycin | | Descriptor: | (3R,4S,5S,7R,9E,11S,12R)-12-ethyl-11-hydroxy-3,5,7,11-tetramethyl-2,8-dioxooxacyclododec-9-en-4-yl 3,4,6-trideoxy-3-(dimethylamino)-beta-D-xylo-hexopyranoside, 23S RIBOSOMAL RNA, 5S RIBOSOMAL RNA | | Authors: | Auerbach, T, Mermershtain, I, Bashan, A, Davidovich, C, Rozenberg, H, Sherman, D.H, Yonath, A. | | Deposit date: | 2009-01-19 | | Release date: | 2010-01-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.71 Å) | | Cite: | Structural basis for the antibacterial activity of the 12-membered-ring mono-sugar macrolide methymycin
Biotechnologia, 1, 2009
|
|
