8DFC
 
 | |
6QMM
 
 | | Crystal structure of Synecochoccus Spermidine Synthase in complex with putrescine, spermidine and MTA | | Descriptor: | (2S)-1-[3-{[(2R)-2-hydroxypropyl]oxy}-2,2-bis({[(2R)-2-hydroxypropyl]oxy}methyl)propoxy]propan-2-ol, 1,4-DIAMINOBUTANE, 5'-DEOXY-5'-METHYLTHIOADENOSINE, ... | | Authors: | Guedez, G, Pothipongsa, A, Incharoensakdi, A, Salminen, T.A. | | Deposit date: | 2019-02-07 | | Release date: | 2019-03-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Crystal structure of dimericSynechococcusspermidine synthase with bound polyamine substrate and product.
Biochem.J., 476, 2019
|
|
6TTN
 
 | | N-terminally truncated hyoscyamine 6-hydroxylase (tH6H) in complex with N-oxalylglycine and hyoscyamine | | Descriptor: | 1,2-ETHANEDIOL, Hyoscyamine 6 beta-hydroxylase, N-OXALYLGLYCINE, ... | | Authors: | Kluza, A, Mrugala, B, Porebski, P.J, Kurpiewska, K, Niedzialkowska, E, Weiss, M.S, Borowski, T. | | Deposit date: | 2019-12-29 | | Release date: | 2020-03-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Regioselectivity of hyoscyamine 6 beta-hydroxylase-catalysed hydroxylation as revealed by high-resolution structural information and QM/MM calculations.
Dalton Trans, 49, 2020
|
|
5WKI
 
 | | Crystal structure of PG90 TCR-CD1b-PG complex | | Descriptor: | (19S,22R,25R)-22,25,26-trihydroxy-16,22-dioxo-17,21,23-trioxa-22lambda~5~-phosphahexacosan-19-yl (9E)-octadec-9-enoate, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Shahine, A, Gras, S, Rossjohn, J. | | Deposit date: | 2017-07-25 | | Release date: | 2017-11-01 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | A molecular basis of human T cell receptor autoreactivity toward self-phospholipids.
Sci Immunol, 2, 2017
|
|
6H8E
 
 | | Truncated derivative of the C-terminal domain of the TssA component of the type VI secretion system from Burkholderia cenocepacia | | Descriptor: | Type VI secretion protein ImpA | | Authors: | Dix, S.R, Owen, H.J, Sun, R, Ahmad, A, Shastri, S, Spiewak, H.L, Mosby, D.J, Harris, M.J, Batters, S.L, Brooker, T.A, Tzokov, S.B, Sedelnikova, S.E, Baker, P.J, Bullough, P.A, Rice, D.W, Thomas, M.S. | | Deposit date: | 2018-08-02 | | Release date: | 2018-11-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural insights into the function of type VI secretion system TssA subunits.
Nat Commun, 9, 2018
|
|
8TBU
 
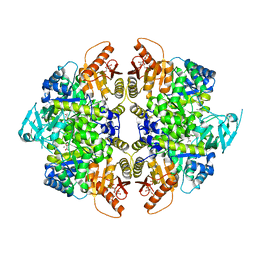 | | Structure of human erythrocyte pyruvate kinase in complex with an allosteric activator Compound 12 | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, 6-[(4-hydroxyphenyl)methyl]-2,4-dimethyl-4,6-dihydro-5H-[1,3]thiazolo[5',4':4,5]pyrrolo[2,3-d]pyridazin-5-one, MANGANESE (II) ION, ... | | Authors: | Jin, L, Padyana, A. | | Deposit date: | 2023-06-29 | | Release date: | 2023-12-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure-Based Design of AG-946, a Pyruvate Kinase Activator.
Chemmedchem, 19, 2024
|
|
8T5F
 
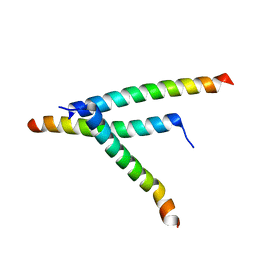 | | De novo design of high-affinity protein binders to bioactive helical peptides | | Descriptor: | Parathyroid hormone | | Authors: | Torres, S.V, Leung, P.J.Y, Bera, A.K, Baker, D, Kang, A. | | Deposit date: | 2023-06-13 | | Release date: | 2024-01-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | De novo design of high-affinity binders of bioactive helical peptides.
Nature, 626, 2024
|
|
8DDL
 
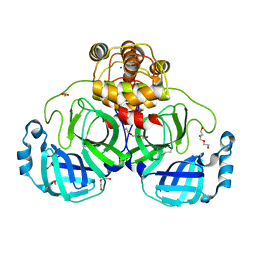 | | SARS-CoV-2 Main Protease (Mpro) H163A Mutant Apo Structure | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, ORF1a polyprotein, ... | | Authors: | Tran, N, McLeod, M.J, Kalyaanamoorthy, S, Ganesan, A, Holyoak, T. | | Deposit date: | 2022-06-18 | | Release date: | 2023-06-28 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The H163A mutation unravels an oxidized conformation of the SARS-CoV-2 main protease.
Nat Commun, 14, 2023
|
|
6WYJ
 
 | | Cryo-EM structure of the GltPh L152C-G321C mutant in the intermediate state | | Descriptor: | ASPARTIC ACID, Glutamate transporter homolog | | Authors: | Font, J, Chen, I, Sobti, M, Stewart, A.G, Ryan, R.M. | | Deposit date: | 2020-05-13 | | Release date: | 2021-02-17 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Glutamate transporters have a chloride channel with two hydrophobic gates.
Nature, 591, 2021
|
|
8TBS
 
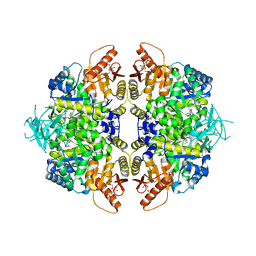 | | Structure of human erythrocyte pyruvate kinase in complex with an allosteric activator AG-946 | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, 6-[(6-aminopyridin-2-yl)methyl]-4-methyl-2-[(1H-pyrazol-3-yl)methyl]-4,6-dihydro-5H-[1,3]thiazolo[5',4':4,5]pyrrolo[2,3-d]pyridazin-5-one, MANGANESE (II) ION, ... | | Authors: | Jin, L, Padyana, A. | | Deposit date: | 2023-06-29 | | Release date: | 2023-12-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure-Based Design of AG-946, a Pyruvate Kinase Activator.
Chemmedchem, 19, 2024
|
|
5WP6
 
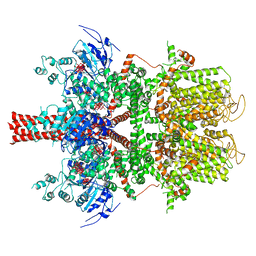 | | Cryo-EM structure of a human TRPM4 channel in complex with calcium and decavanadate | | Descriptor: | DECAVANADATE, Transient receptor potential cation channel subfamily M member 4 | | Authors: | Winkler, P.A, Huang, Y, Sun, W, Du, J, Lu, W. | | Deposit date: | 2017-08-03 | | Release date: | 2017-12-13 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Electron cryo-microscopy structure of a human TRPM4 channel.
Nature, 552, 2017
|
|
6OGJ
 
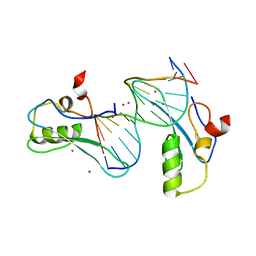 | | MeCP2 MBD in complex with DNA | | Descriptor: | DNA (5'-D(*CP*GP*GP*AP*GP*TP*GP*TP*AP*GP*GP*C)-3'), DNA (5'-D(*GP*CP*CP*TP*AP*CP*AP*CP*TP*CP*CP*G)-3'), Methyl-CpG-binding protein 2, ... | | Authors: | Lei, M, Tempel, W, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-04-02 | | Release date: | 2019-05-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Plasticity at the DNA recognition site of the MeCP2 mCG-binding domain.
Biochim Biophys Acta Gene Regul Mech, 1862, 2019
|
|
8DFD
 
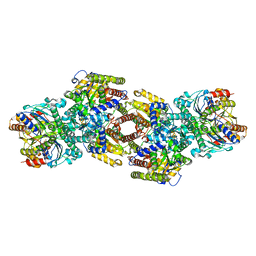 | |
5F13
 
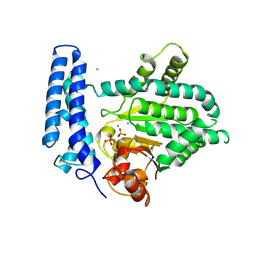 | | Structure of Mn bound DUF89 from Saccharomyces cerevisiae | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, MANGANESE (II) ION, ... | | Authors: | Nocek, B, Skarina, T, Joachimiak, A, Savchenko, A, Yakunin, A. | | Deposit date: | 2015-11-30 | | Release date: | 2016-03-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.393 Å) | | Cite: | A family of metal-dependent phosphatases implicated in metabolite damage-control.
Nat.Chem.Biol., 12, 2016
|
|
7QT9
 
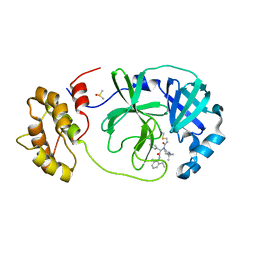 | | Room temperature In-situ SARS-CoV-2 MPRO with bound Z4439011584 | | Descriptor: | DIMETHYL SULFOXIDE, N-(5-tert-butyl-1H-pyrazol-3-yl)-N-[(1R)-2-[(2-ethyl-6-methylphenyl)amino]-2-oxo-1-(pyridin-3-yl)ethyl]propanamide, Non-structural protein 6 | | Authors: | Horrell, S, Gildae, R.J, Axford, D, Owen, C.D, Lukacik, P, Strain-Damerell, C, Owen, R.L, Walsh, M.A. | | Deposit date: | 2022-01-14 | | Release date: | 2022-05-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | xia2.multiplex: a multi-crystal data-analysis pipeline.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
4QPF
 
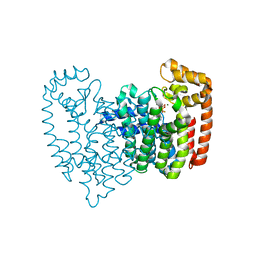 | | New lower bone affinity bisphosphonate drug design for effective use in diseases characterized by abnormal bone resorption | | Descriptor: | Farnesyl pyrophosphate synthase, MAGNESIUM ION, [1-fluoro-2-(imidazo[1,2-a]pyridin-3-yl)ethane-1,1-diyl]bis(phosphonic acid) | | Authors: | Ebetino, F.H, Lundy, M, Kwaasi, A.A, Dunford, J.E, Duan, Z, Triffitt, J, Mazur, A, Jeans, G, Barnett, B.L, Russell, R.G.G. | | Deposit date: | 2014-06-23 | | Release date: | 2015-06-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | New lower bone affinity bisphosphonate drug design for effective use in diseases characterized by abnormal bone resorption
To be Published
|
|
8Q3U
 
 | | Crystal structure of a fentanyl derivative in complex with human CA VII | | Descriptor: | Carbonic anhydrase 7, GLYCEROL, ZINC ION, ... | | Authors: | Alterio, V, Di Fiore, A, De Simone, G. | | Deposit date: | 2023-08-04 | | Release date: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Discovery of a novel series of potent carbonic anhydrase inhibitors with selective affinity for mu Opioid receptor for Safer and long-lasting analgesia.
Eur.J.Med.Chem., 260, 2023
|
|
7T5O
 
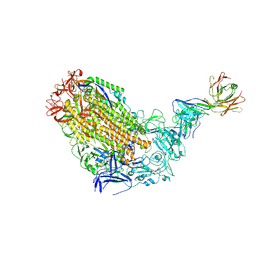 | | VFLIP Spike Trimer with GAR03 | | Descriptor: | GAR03 Fab heavy chain, GAR03 Fab light chain, Spike glycoprotein | | Authors: | Sobti, M, Stewart, A.G, Rouet, R, Langley, D.B. | | Deposit date: | 2021-12-12 | | Release date: | 2023-01-25 | | Last modified: | 2023-08-09 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | Broadly neutralizing SARS-CoV-2 antibodies through epitope-based selection from convalescent patients.
Nat Commun, 14, 2023
|
|
8T5E
 
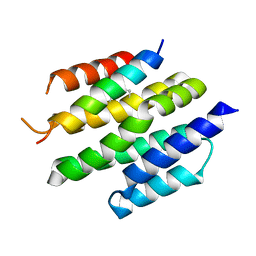 | | De novo design of high-affinity protein binders to bioactive helical peptides | | Descriptor: | Bcl-2-like protein 11, Bim_fulldiff | | Authors: | Torres, S.V, Leung, P.J.Y, Bera, A.K, Baker, D, Kang, A. | | Deposit date: | 2023-06-13 | | Release date: | 2024-01-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | De novo design of high-affinity binders of bioactive helical peptides.
Nature, 626, 2024
|
|
6HCN
 
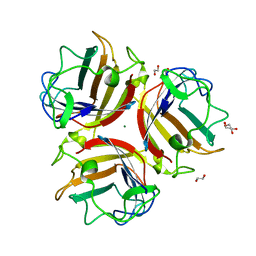 | | Adenovirus Type 5 Fiber Knob protein at 1.49A resolution | | Descriptor: | 1,2-ETHANEDIOL, Fiber protein, MAGNESIUM ION | | Authors: | Rizkallah, P.J, Parker, A.L, Baker, A.T. | | Deposit date: | 2018-08-15 | | Release date: | 2019-02-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Diversity within the adenovirus fiber knob hypervariable loops influences primary receptor interactions.
Nat Commun, 10, 2019
|
|
5IBZ
 
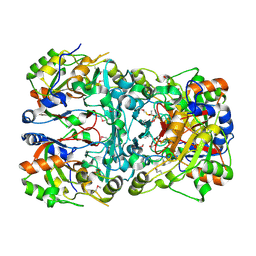 | | Crystal structure of a novel cyclase (pfam04199). | | Descriptor: | ACETYLPHOSPHATE, TRIETHYLENE GLYCOL, Uncharacterized protein | | Authors: | Nocek, B, Skarina, T, Brown, G, Joachimiak, A, Savchenko, A, Yakunin, A. | | Deposit date: | 2016-02-22 | | Release date: | 2017-08-09 | | Method: | X-RAY DIFFRACTION (1.611 Å) | | Cite: | Crystal structure of a novel cyclase (pfam04199).
To Be Published
|
|
6WYK
 
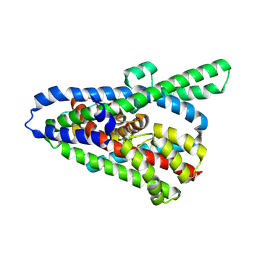 | | Cryo-EM structure of the GltPh L152C-G321C mutant in the intermediate chloride conducting state. | | Descriptor: | ASPARTIC ACID, Glutamate transporter homolog | | Authors: | Font, J, Chen, I, Sobti, M, Stewart, A.G, Ryan, R.M. | | Deposit date: | 2020-05-13 | | Release date: | 2021-02-17 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Glutamate transporters have a chloride channel with two hydrophobic gates.
Nature, 591, 2021
|
|
8T7E
 
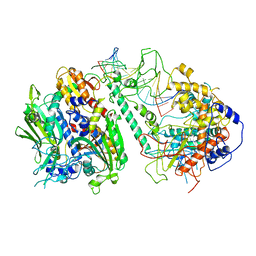 | | Cryo-EM structure of the Backtracking Initiation Complex (VII) of Human Mitochondrial DNA Polymerase Gamma | | Descriptor: | DNA polymerase subunit gamma-1, DNA polymerase subunit gamma-2, mitochondrial, ... | | Authors: | Nayak, A.R, Buchel, G, Herbine, K.H, Sarfallah, A, Sokolova, V.O, Zamudio-Ochoa, A, Temiakov, D. | | Deposit date: | 2023-06-20 | | Release date: | 2024-01-10 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Structural basis for DNA proofreading.
Nat Commun, 14, 2023
|
|
7QT6
 
 | | Room temperature In-situ SARS-CoV-2 MPRO with bound Z1367324110 | | Descriptor: | 1-methyl-3,4-dihydro-2~{H}-quinoline-7-sulfonamide, 3C-like proteinase nsp5, DIMETHYL SULFOXIDE | | Authors: | Horrell, S, Gildae, R.J, Axford, D, Owen, C.D, Lukacik, P, Strain-Damerell, C, Owen, R.L, Walsh, M.A. | | Deposit date: | 2022-01-14 | | Release date: | 2022-05-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | xia2.multiplex: a multi-crystal data-analysis pipeline.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7QT5
 
 | | Room temperature In-situ SARS-CoV-2 MPRO with bound Z31792168 | | Descriptor: | 2-cyclohexyl-~{N}-pyridin-3-yl-ethanamide, 3C-like proteinase nsp5, DIMETHYL SULFOXIDE | | Authors: | Horrell, S, Gildae, R.J, Axford, D, Owen, C.D, Lukacik, P, Strain-Damerell, C, Owen, R.L, Walsh, M.A. | | Deposit date: | 2022-01-14 | | Release date: | 2022-05-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | xia2.multiplex: a multi-crystal data-analysis pipeline.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
