5HSM
 
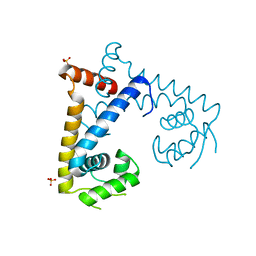 | | CRYSTAL STRUCTURE OF MYCOBACTERIUM TUBERCULOSIS MARR FAMILY PROTEIN RV2887 | | Descriptor: | SULFATE ION, Uncharacterized HTH-type transcriptional regulator Rv2887 | | Authors: | Gao, Y.R, Li, D.F, Wang, D.C, Bi, L.J. | | Deposit date: | 2016-01-26 | | Release date: | 2017-02-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural analysis of the regulatory mechanism of MarR protein Rv2887 in M. tuberculosis
Sci Rep, 7, 2017
|
|
8TZN
 
 | |
8TZW
 
 | |
8U08
 
 | |
5T0L
 
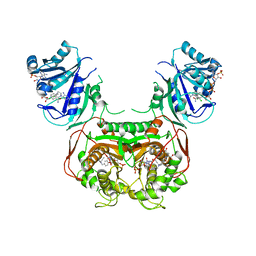 | | Crystal structure of Toxoplasma gondii TS-DHFR complexed with NADPH, dUMP, PDDF and 5-(4-(3,4-dichlorophenyl)piperazin-1-yl)pyrimidine-2,4-diamine (TRC-15) | | Descriptor: | 10-PROPARGYL-5,8-DIDEAZAFOLIC ACID, 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, 5-[4-(3,4-dichlorophenyl)piperazin-1-yl]pyrimidine-2,4-diamine, ... | | Authors: | Thomas, S.B, Chen, Z, Lu, H, Li, Y. | | Deposit date: | 2016-08-16 | | Release date: | 2016-09-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.13 Å) | | Cite: | Discovery of Potent and Selective Leads against Toxoplasma gondii Dihydrofolate Reductase via Structure-Based Design.
ACS Med Chem Lett, 7, 2016
|
|
8GOU
 
 | | Omicron BA.4/5 SARS-CoV-2 S in complex with TH003 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, TH003 Fab heavy chain, ... | | Authors: | Guo, Y, Zhang, G, Liang, J, Liu, F, Rao, Z. | | Deposit date: | 2022-08-25 | | Release date: | 2023-06-28 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Discovery and characterization of potent pan-variant SARS-CoV-2 neutralizing antibodies from individuals with Omicron breakthrough infection.
Nat Commun, 14, 2023
|
|
8GPY
 
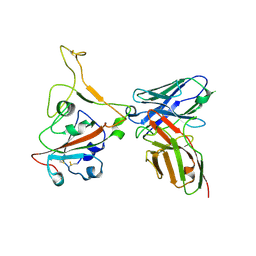 | | Crystal structure of Omicron BA.4/5 RBD in complex with a neutralizing antibody scFv | | Descriptor: | Spike protein S1, scFv | | Authors: | Gao, Y.X, Song, Z.D, Wang, W.M, Guo, Y. | | Deposit date: | 2022-08-27 | | Release date: | 2023-06-28 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Discovery and characterization of potent pan-variant SARS-CoV-2 neutralizing antibodies from individuals with Omicron breakthrough infection.
Nat Commun, 14, 2023
|
|
4JV7
 
 | | Co-crystal structure of MDM2 with inhibitor (2S,5R,6S)-2-benzyl-5,6-bis(4-bromophenyl)-4-methylmorpholin-3-one | | Descriptor: | (2S,5R,6S)-2-benzyl-5,6-bis(4-bromophenyl)-4-methylmorpholin-3-one, E3 ubiquitin-protein ligase Mdm2, SULFATE ION | | Authors: | Huang, X, Gonzalez-Lopez de Turiso, F, Sun, D, Yosup, R, Bartberger, M.D, Beck, H.P, Cannon, J, Shaffer, P, Oliner, J.D, Olson, S.H, Medina, J.C. | | Deposit date: | 2013-03-25 | | Release date: | 2013-05-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Rational Design and Binding Mode Duality of MDM2-p53 Inhibitors.
J.Med.Chem., 56, 2013
|
|
4JVR
 
 | | Co-crystal structure of MDM2 with inhibitor (2'S,3R,4'S,5'R)-N-(2-aminoethyl)-6-chloro-4'-(3-chloro-2-fluorophenyl)-2'-(2,2-dimethylpropyl)-2-oxo-1,2-dihydrospiro[indole-3,3'-pyrrolidine]-5'-carboxamide | | Descriptor: | (2'S,3R,4'S,5'R)-N-(2-aminoethyl)-6-chloro-4'-(3-chloro-2-fluorophenyl)-2'-(2,2-dimethylpropyl)-2-oxo-1,2-dihydrospiro[indole-3,3'-pyrrolidine]-5'-carboxamide, E3 ubiquitin-protein ligase Mdm2 | | Authors: | Huang, X, Gonzalez-Lopez de Turiso, F, Sun, D, Yosup, R, Bartberger, M.D, Beck, H.P, Cannon, J, Shaffer, P, Oliner, J.D, Olson, S.H, Medina, J.C. | | Deposit date: | 2013-03-26 | | Release date: | 2013-05-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Rational Design and Binding Mode Duality of MDM2-p53 Inhibitors.
J.Med.Chem., 56, 2013
|
|
4JVE
 
 | | Co-crystal structure of MDM2 with inhibitor (2R,3E)-2-[(2S,3R,6S)-2,3-bis(4-chlorophenyl)-6-(4-fluorobenzyl)-5-oxomorpholin-4-yl]pent-3-enoic acid | | Descriptor: | (2R,3E)-2-[(2S,3R,6S)-2,3-bis(4-chlorophenyl)-6-(4-fluorobenzyl)-5-oxomorpholin-4-yl]pent-3-enoic acid, E3 ubiquitin-protein ligase Mdm2 | | Authors: | Huang, X, Gonzalez-Lopez de Turiso, F, Sun, D, Yosup, R, Bartberger, M.D, Beck, H.P, Cannon, J, Shaffer, P, Oliner, J.D, Olson, S.H, Medina, J.C. | | Deposit date: | 2013-03-25 | | Release date: | 2013-05-01 | | Last modified: | 2013-06-05 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Rational Design and Binding Mode Duality of MDM2-p53 Inhibitors.
J.Med.Chem., 56, 2013
|
|
8GT6
 
 | | human STING With agonist HB3089 | | Descriptor: | 1-[(2E)-4-{5-carbamoyl-2-[(1-ethyl-3-methyl-1H-pyrazole-5-carbonyl)amino]-7-[3-(morpholin-4-yl)propoxy]-1H-benzimidazol-1-yl}but-2-en-1-yl]-2-[(1-ethyl-3-methyl-1H-pyrazole-5-carbonyl)amino]-7-methyl-1H-furo[3,2-e]benzimidazole-5-carboxamide, Stimulator of interferon genes protein | | Authors: | Wang, Z, Yu, X. | | Deposit date: | 2022-09-07 | | Release date: | 2022-12-28 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Structural insights into a shared mechanism of human STING activation by a potent agonist and an autoimmune disease-associated mutation.
Cell Discov, 8, 2022
|
|
8GSZ
 
 | | Structure of STING SAVI-related mutant V147L | | Descriptor: | Stimulator of interferon genes protein | | Authors: | Wang, Z, Yu, X. | | Deposit date: | 2022-09-07 | | Release date: | 2022-12-28 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.65 Å) | | Cite: | Structural insights into a shared mechanism of human STING activation by a potent agonist and an autoimmune disease-associated mutation.
Cell Discov, 8, 2022
|
|
8KCO
 
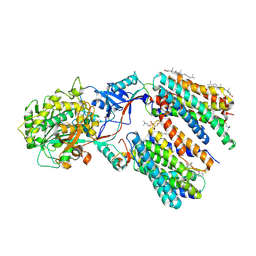 | | Cryo-EM structure of human gamma-secretase in complex with RO4929097 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2,2-dimethyl-N-[(7S)-6-oxo-5,7-dihydrobenzo[d][1]benzazepin-7-yl]-N'-(2,2,3,3,3-pentafluoropropyl)propanediamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Guo, X, Li, H, Kai, U, Yan, C, Lei, J, Zhou, R, Shi, Y. | | Deposit date: | 2023-08-08 | | Release date: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | CryoEM structure of PS1-containing gamma-secretase in complex with MRK-560
To Be Published
|
|
4JV9
 
 | | Co-crystal structure of MDM2 with inhibitor (2S,5R,6S)-2-benzyl-5,6-bis(4-chlorophenyl)-4-methylmorpholin-3-one | | Descriptor: | (2S,5R,6S)-2-benzyl-5,6-bis(4-chlorophenyl)-4-methylmorpholin-3-one, E3 ubiquitin-protein ligase Mdm2, SULFATE ION | | Authors: | Huang, X, Gonzalez-Lopez de Turiso, F, Sun, D, Yosup, R, Bartberger, M.D, Beck, H.P, Cannon, J, Shaffer, P, Oliner, J.D, Olson, S.H, Medina, J.C. | | Deposit date: | 2013-03-25 | | Release date: | 2013-05-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Rational Design and Binding Mode Duality of MDM2-p53 Inhibitors.
J.Med.Chem., 56, 2013
|
|
6SCY
 
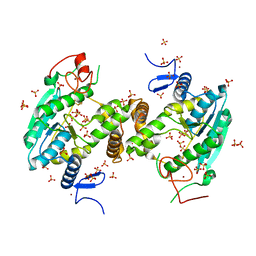 | | U34-tRNA thiolase NcsA from Methanococcus maripaludis with its [4Fe-4S] cluster | | Descriptor: | IRON/SULFUR CLUSTER, SULFATE ION, ZINC ION, ... | | Authors: | Bimai, O, Legrand, P, Golinelli-Pimpaneau, B. | | Deposit date: | 2019-07-25 | | Release date: | 2020-08-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | The thiolation of uridine 34 in tRNA, which controls protein translation, depends on a [4Fe-4S] cluster in the archaeum Methanococcus maripaludis.
Sci Rep, 13, 2023
|
|
3OBT
 
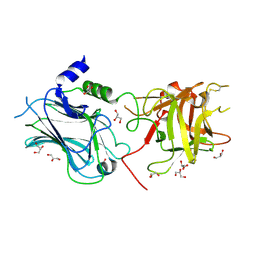 | |
6UWU
 
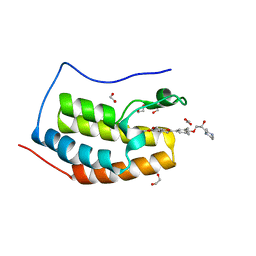 | | Crystal Structure of the first bromodomain of human BRD4 in complex with the inhibitor ZL0516 | | Descriptor: | 1,2-ETHANEDIOL, 2-{4-[(2R)-2-hydroxy-3-(4-methylpiperazin-1-yl)propoxy]-3,5-dimethylphenyl}-5,7-dimethoxy-4H-1-benzopyran-4-one, Bromodomain-containing protein 4 | | Authors: | Leonard, P.G, Joseph, S. | | Deposit date: | 2019-11-05 | | Release date: | 2020-04-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of Orally Bioavailable Chromone Derivatives as Potent and Selective BRD4 Inhibitors: Scaffold Hopping, Optimization, and Pharmacological Evaluation.
J.Med.Chem., 63, 2020
|
|
3OBR
 
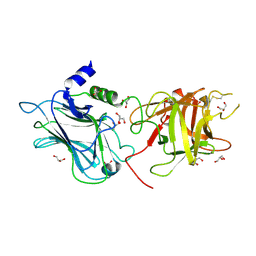 | |
4JWR
 
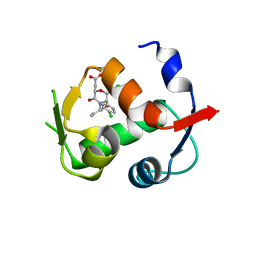 | |
4HBM
 
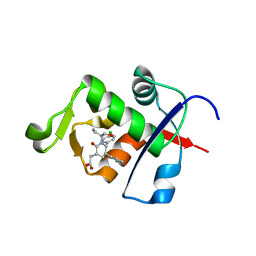 | | Ordering of the N Terminus of Human MDM2 by Small Molecule Inhibitors | | Descriptor: | E3 ubiquitin-protein ligase Mdm2, {(3R,5R,6S)-5-(3-chlorophenyl)-6-(4-chlorophenyl)-1-[(2S)-1-hydroxybutan-2-yl]-2-oxopiperidin-3-yl}acetic acid | | Authors: | Huang, X. | | Deposit date: | 2012-09-28 | | Release date: | 2012-10-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Ordering of the N-terminus of human MDM2 by small molecule inhibitors.
J.Am.Chem.Soc., 134, 2012
|
|
1Y4K
 
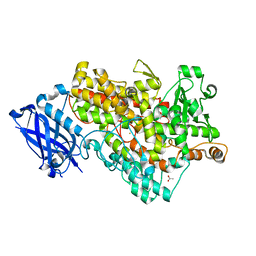 | | Lipoxygenase-1 (Soybean) at 100K, N694G Mutant | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, FE (II) ION, ... | | Authors: | Chruszcz, M, Segraves, E, Holman, T.R, Minor, W. | | Deposit date: | 2004-12-01 | | Release date: | 2005-12-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Kinetic, spectroscopic, and structural investigations of the soybean lipoxygenase-1 first-coordination sphere mutant, Asn694Gly.
Biochemistry, 45, 2006
|
|
3NMZ
 
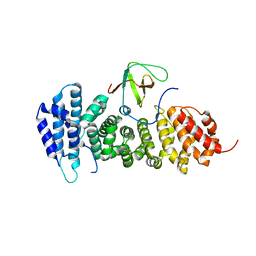 | | Crystal structure of APC complexed with Asef | | Descriptor: | APC variant protein, Rho guanine nucleotide exchange factor 4 | | Authors: | Zhang, Z, Chen, L, Gao, L, Lin, K, Wu, G. | | Deposit date: | 2010-06-23 | | Release date: | 2011-07-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Structural basis for the recognition of Asef by adenomatous polyposis coli.
Cell Res., 22, 2012
|
|
3NMX
 
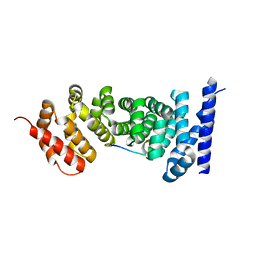 | | Crystal structure of APC complexed with Asef | | Descriptor: | APC variant protein, Rho guanine nucleotide exchange factor 4 | | Authors: | Zhang, Z, Chen, L, Gao, L, Lin, K, Wu, G. | | Deposit date: | 2010-06-22 | | Release date: | 2011-07-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for the recognition of Asef by adenomatous polyposis coli.
Cell Res., 22, 2012
|
|
5ULA
 
 | |
3NMW
 
 | | Crystal structure of armadillo repeats domain of APC | | Descriptor: | APC variant protein, SULFATE ION | | Authors: | Zhang, Z, Chen, L, Gao, L, Lin, K, Wu, G. | | Deposit date: | 2010-06-22 | | Release date: | 2011-07-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis for the recognition of Asef by adenomatous polyposis coli.
Cell Res., 22, 2012
|
|
