6PAX
 
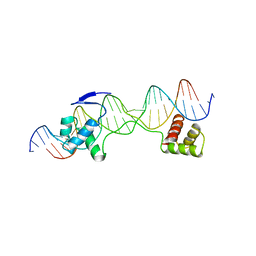 | | CRYSTAL STRUCTURE OF THE HUMAN PAX-6 PAIRED DOMAIN-DNA COMPLEX REVEALS A GENERAL MODEL FOR PAX PROTEIN-DNA INTERACTIONS | | Descriptor: | 26 NUCLEOTIDE DNA, HOMEOBOX PROTEIN PAX-6 | | Authors: | Xu, H.E, Rould, M.A, Xu, W, Epstein, J.A, Maas, R.L, Pabo, C.O. | | Deposit date: | 1999-04-22 | | Release date: | 1999-07-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the human Pax6 paired domain-DNA complex reveals specific roles for the linker region and carboxy-terminal subdomain in DNA binding.
Genes Dev., 13, 1999
|
|
1KKQ
 
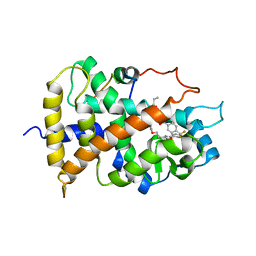 | | Crystal structure of the human PPAR-alpha ligand-binding domain in complex with an antagonist GW6471 and a SMRT corepressor motif | | Descriptor: | N-((2S)-2-({(1Z)-1-METHYL-3-OXO-3-[4-(TRIFLUOROMETHYL) PHENYL]PROP-1-ENYL}AMINO)-3-{4-[2-(5-METHYL-2-PHENYL-1,3-OXAZOL-4-YL)ETHOXY]PHENYL}PROPYL)PROPANAMIDE, NUCLEAR RECEPTOR CO-REPRESSOR 2, PEROXISOME PROLIFERATOR ACTIVATED RECEPTOR | | Authors: | Xu, H.E, Stanley, T.B, Montana, V.G, Lambert, M.H, Shearer, B.G, Cobb, J.E, McKee, D.D, Galardi, C.M, Nolte, R.T, Parks, D.J. | | Deposit date: | 2001-12-10 | | Release date: | 2002-02-20 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for antagonist-mediated recruitment of nuclear co-repressors by PPARalpha.
Nature, 415, 2002
|
|
1GWX
 
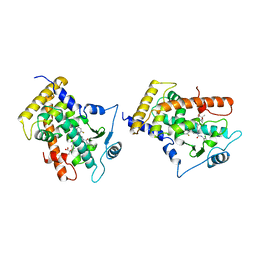 | | MOLECULAR RECOGNITION OF FATTY ACIDS BY PEROXISOME PROLIFERATOR-ACTIVATED RECEPTORS | | Descriptor: | 2-(4-{3-[1-[2-(2-CHLORO-6-FLUORO-PHENYL)-ETHYL]-3-(2,3-DICHLORO-PHENYL)-UREIDO]-PROPYL}-PHENOXY)-2-METHYL-PROPIONIC ACID, PROTEIN (PPAR-DELTA) | | Authors: | Xu, H.E, Lambert, M.H, Montana, V.G, Park, D.J, Blanchard, S, Brown, P, Sternbach, D, Lehmann, J, Bruce, G.W, Willson, T.M, Kliewer, S.A, Milburn, M.V. | | Deposit date: | 1999-03-17 | | Release date: | 2000-03-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular recognition of fatty acids by peroxisome proliferator-activated receptors.
Mol.Cell, 3, 1999
|
|
1K74
 
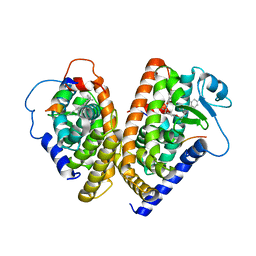 | | The 2.3 Angstrom resolution crystal structure of the heterodimer of the human PPARgamma and RXRalpha ligand binding domains respectively bound with GW409544 and 9-cis retinoic acid and co-activator peptides. | | Descriptor: | (9cis)-retinoic acid, 2-(1-METHYL-3-OXO-3-PHENYL-PROPYLAMINO)-3-{4-[2-(5-METHYL-2-PHENYL-OXAZOL-4-YL)-ETHOXY]-PHENYL}-PROPIONIC ACID, Peroxisome proliferator activated receptor gamma, ... | | Authors: | Xu, H.E, Lambert, M.H, Montana, V.G, Moore, L.B, Collins, J.L, Oplinger, J.A, Kliewer, S.A, Gampe Jr, R.T, McKee, D.D, Moore, J.T, Willson, T.M. | | Deposit date: | 2001-10-18 | | Release date: | 2001-12-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural determinants of ligand binding selectivity between the peroxisome proliferator-activated receptors.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1K7L
 
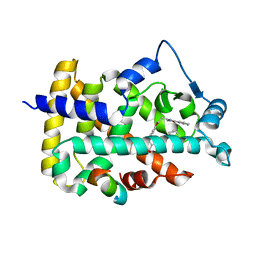 | | The 2.5 Angstrom resolution crystal structure of the human PPARalpha ligand binding domain bound with GW409544 and a co-activator peptide. | | Descriptor: | 2-(1-METHYL-3-OXO-3-PHENYL-PROPYLAMINO)-3-{4-[2-(5-METHYL-2-PHENYL-OXAZOL-4-YL)-ETHOXY]-PHENYL}-PROPIONIC ACID, Peroxisome proliferator activated receptor alpha, YTTRIUM (III) ION, ... | | Authors: | Xu, H.E, Lambert, M.H, Montana, V.G, Moore, L.B, Collins, J.L, Oplinger, J.A, Kliewer, S.A, Gampe Jr, R.T, McKee, D.D, Moore, J.T, Willson, T.M. | | Deposit date: | 2001-10-19 | | Release date: | 2001-12-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural determinants of ligand binding selectivity between the peroxisome proliferator-activated receptors.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
3GWX
 
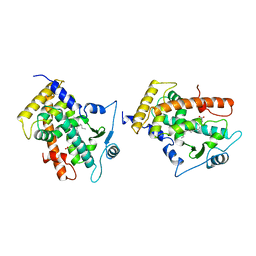 | | MOLECULAR RECOGNITION OF FATTY ACIDS BY PEROXISOME PROLIFERATOR-ACTIVATED RECEPTORS | | Descriptor: | 5,8,11,14,17-EICOSAPENTAENOIC ACID, PROTEIN (PEROXISOME PROLIFERATOR ACTIVATED RECEPTOR (PPAR-DELTA)) | | Authors: | Xu, H.E, Lambert, M.H, Montana, V.G, Parks, D.J, Blanchard, S.G, Brown, P.J, Sternbach, D.D, Lehmann, J.M, Wisely, G.B, Willson, T.M, Kliewer, S.A, Milburn, M.V. | | Deposit date: | 1999-04-26 | | Release date: | 2000-04-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular recognition of fatty acids by peroxisome proliferator-activated receptors.
Mol.Cell, 3, 1999
|
|
2GWX
 
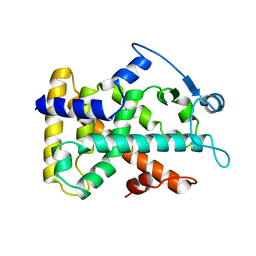 | | MOLECULAR RECOGNITION OF FATTY ACIDS BY PEROXISOME PROLIFERATOR-ACTIVATED RECEPTORS | | Descriptor: | PROTEIN (PPAR-DELTA) | | Authors: | Xu, H.E, Lambert, M.H, Montana, V.G, Park, D.J, Blanchard, S, Brown, P, Sternbach, D, Lehmann, J, Bruce, G.W, Willson, T.M, Kliewer, S.A, Milburn, M.V. | | Deposit date: | 1999-03-11 | | Release date: | 2000-03-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular recognition of fatty acids by peroxisome proliferator-activated receptors.
Mol.Cell, 3, 1999
|
|
3BQD
 
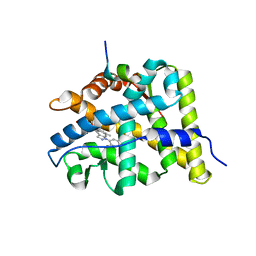 | | Doubling the Size of the Glucocorticoid Receptor Ligand Binding Pocket by Deacylcortivazol | | Descriptor: | 1-[(1R,2R,3aS,3bS,10aR,10bS,11S,12aS)-1,11-dihydroxy-2,5,10a,12a-tetramethyl-7-phenyl-1,2,3,3a,3b,7,10,10a,10b,11,12,12a-dodecahydrocyclopenta[5,6]naphtho[1,2-f]indazol-1-yl]-2-hydroxyethanone, Glucocorticoid receptor, Nuclear receptor coactivator 1 | | Authors: | Xu, H.E. | | Deposit date: | 2007-12-20 | | Release date: | 2008-01-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Doubling the size of the glucocorticoid receptor ligand binding pocket by deacylcortivazol.
Mol.Cell.Biol., 28, 2008
|
|
5JGC
 
 | | Crystal structure of the rice Topless related protein 2 (TPR2) N-terminal topless domain (1-209) L111A, L130A, L179A and I195A mutant | | Descriptor: | Protein TPR1, ZINC ION | | Authors: | Ke, J, Ma, H, Gu, X, Brunzelle, J.S, Xu, H.E, Melcher, K. | | Deposit date: | 2016-04-20 | | Release date: | 2017-07-05 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | A D53 repression motif induces oligomerization of TOPLESS corepressors and promotes assembly of a corepressor-nucleosome complex.
Sci Adv, 3, 2017
|
|
6VN7
 
 | | Cryo-EM structure of an activated VIP1 receptor-G protein complex | | Descriptor: | CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Duan, J, Shen, D.-D, Zhou, X.E, Liu, Q.-F, Zhuang, Y.-W, Zhang, H.-B, Xu, P.-Y, Ma, S.-S, He, X.-H, Melcher, K, Zhang, Y, Xu, H.E, Yi, J. | | Deposit date: | 2020-01-29 | | Release date: | 2020-09-02 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structure of an activated VIP1 receptor-G protein complex revealed by a NanoBiT tethering strategy.
Nat Commun, 11, 2020
|
|
6UH3
 
 | | Crystal structure of bacterial heliorhodopsin 48C12 | | Descriptor: | DI(HYDROXYETHYL)ETHER, Heliorhodopsin, PALMITIC ACID, ... | | Authors: | Lu, Y, Zhou, X.E, Gao, X, Xia, R, Xu, Z, Wang, N, Leng, Y, Melcher, K, Xu, H.E, He, Y. | | Deposit date: | 2019-09-26 | | Release date: | 2019-12-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of heliorhodopsin 48C12.
Cell Res., 30, 2020
|
|
7BV1
 
 | | Cryo-EM structure of the apo nsp12-nsp7-nsp8 complex | | Descriptor: | Non-structural protein 7, Non-structural protein 8, RNA-directed RNA polymerase, ... | | Authors: | Yin, W, Mao, C, Luan, X, Shen, D, Shen, Q, Su, H, Wang, X, Zhou, F, Zhao, W, Gao, M, Chang, S, Xie, Y.C, Tian, G, Jiang, H.W, Tao, S.C, Shen, J, Jiang, Y, Jiang, H, Xu, Y, Zhang, S, Zhang, Y, Xu, H.E. | | Deposit date: | 2020-04-09 | | Release date: | 2020-04-22 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis for inhibition of the RNA-dependent RNA polymerase from SARS-CoV-2 by remdesivir.
Science, 368, 2020
|
|
7BV2
 
 | | The nsp12-nsp7-nsp8 complex bound to the template-primer RNA and triphosphate form of Remdesivir(RTP) | | Descriptor: | MAGNESIUM ION, Non-structural protein 7, Non-structural protein 8, ... | | Authors: | Yin, W, Mao, C, Luan, X, Shen, D, Shen, Q, Su, H, Wang, X, Zhou, F, Zhao, W, Gao, M, Chang, S, Xie, Y.C, Tian, G, Jiang, H.W, Tao, S.C, Shen, J, Jiang, Y, Jiang, H, Xu, Y, Zhang, S, Zhang, Y, Xu, H.E. | | Deposit date: | 2020-04-09 | | Release date: | 2020-04-22 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Structural basis for inhibition of the RNA-dependent RNA polymerase from SARS-CoV-2 by remdesivir.
Science, 368, 2020
|
|
6PA7
 
 | | The cryo-EM structure of the human DNMT3A2-DNMT3B3 complex bound to nucleosome. | | Descriptor: | CHLORIDE ION, DNA (167-MER), DNA (cytosine-5)-methyltransferase 3A, ... | | Authors: | Xu, T.H, Liu, M, Zhou, X.E, Liang, G, Zhao, G, Xu, H.E, Melcher, K, Jones, P.A. | | Deposit date: | 2019-06-11 | | Release date: | 2020-06-17 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.94 Å) | | Cite: | Structure of nucleosome-bound DNA methyltransferases DNMT3A and DNMT3B.
Nature, 586, 2020
|
|
6PT0
 
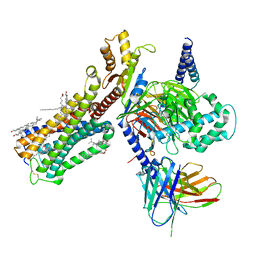 | | Cryo-EM structure of human cannabinoid receptor 2-Gi protein in complex with agonist WIN 55,212-2 | | Descriptor: | CHOLESTEROL, Cannabinoid receptor 2, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Xu, T.H, Xing, C, Zhuang, Y, Feng, Z, Zhou, X.E, Chen, M, Wang, L, Meng, X, Xue, Y, Wang, J, Liu, H, McGuire, T, Zhao, G, Melcher, K, Zhang, C, Xu, H.E, Xie, X.Q. | | Deposit date: | 2019-07-14 | | Release date: | 2020-02-12 | | Last modified: | 2020-03-04 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM Structure of the Human Cannabinoid Receptor CB2-GiSignaling Complex.
Cell, 180, 2020
|
|
4IH1
 
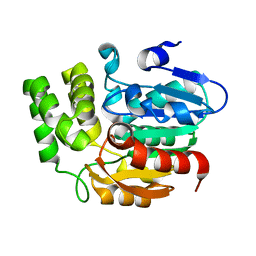 | | Crystal structure of Karrikin Insensitive 2 (KAI2) from Arabidopsis thaliana | | Descriptor: | Hydrolase, alpha/beta fold family protein | | Authors: | Zhou, X.E, Zhao, L.-H, Wu, Z.-S, Yi, W, Li, S, Li, Y, Xu, Y, Xu, T.-H, Liu, Y, Chen, R.-Z, Kovach, A, Kang, Y, Hou, L, He, Y, Zhang, C, Melcher, K, Xu, H.E. | | Deposit date: | 2012-12-18 | | Release date: | 2013-01-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structures of two phytohormone signal-transducing alpha / beta hydrolases: karrikin-signaling KAI2 and strigolactone-signaling DWARF14.
Cell Res., 23, 2013
|
|
4IH9
 
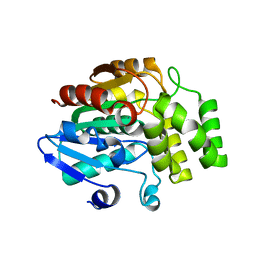 | | Crystal structure of rice DWARF14 (D14) | | Descriptor: | Dwarf 88 esterase | | Authors: | Zhou, X.E, Zhao, L.-H, Wu, Z.-S, Yi, W, Li, S, Li, Y, Xu, Y, Xu, T.-H, Liu, Y, Chen, R.-Z, Kovach, A, Kang, Y, Hou, L, He, Y, Zhang, C, Melcher, K, Xu, H.E. | | Deposit date: | 2012-12-18 | | Release date: | 2013-01-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structures of two phytohormone signal-transducing alpha / beta hydrolases: karrikin-signaling KAI2 and strigolactone-signaling DWARF14.
Cell Res., 23, 2013
|
|
1G1U
 
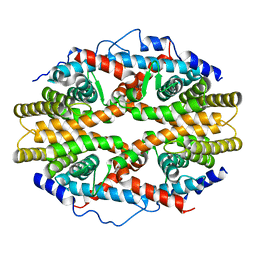 | | THE 2.5 ANGSTROM RESOLUTION CRYSTAL STRUCTURE OF THE RXRALPHA LIGAND BINDING DOMAIN IN TETRAMER IN THE ABSENCE OF LIGAND | | Descriptor: | RETINOIC ACID RECEPTOR RXR-ALPHA | | Authors: | Gampe Jr, R.T, Montana, V.G, Lambert, M.H, Wisely, G.B, Milburn, M.V, Xu, H.E. | | Deposit date: | 2000-10-13 | | Release date: | 2001-04-25 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for autorepression of retinoid X receptor by tetramer formation and the AF-2 helix.
Genes Dev., 14, 2000
|
|
1G5Y
 
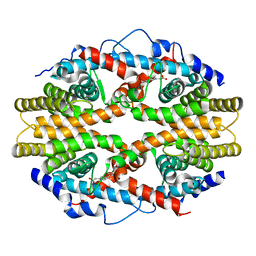 | | THE 2.0 ANGSTROM RESOLUTION CRYSTAL STRUCTURE OF THE RXRALPHA LIGAND BINDING DOMAIN TETRAMER IN THE PRESENCE OF A NON-ACTIVATING RETINOIC ACID ISOMER. | | Descriptor: | RETINOIC ACID, RETINOIC ACID RECEPTOR RXR-ALPHA | | Authors: | Gampe Jr, R.T, Montana, V.G, Lambert, M.H, Wisely, G.B, Milburn, M.V, Xu, H.E. | | Deposit date: | 2000-11-02 | | Release date: | 2001-05-02 | | Last modified: | 2018-04-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for autorepression of retinoid X receptor by tetramer formation and the AF-2 helix.
Genes Dev., 14, 2000
|
|
1GCB
 
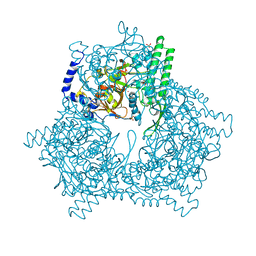 | | GAL6, YEAST BLEOMYCIN HYDROLASE DNA-BINDING PROTEASE (THIOL) | | Descriptor: | GAL6 HG (EMTS) DERIVATIVE, GLYCEROL, MERCURY (II) ION, ... | | Authors: | Joshua-Tor, L, Xu, H.E, Johnston, S.A, Rees, D.C. | | Deposit date: | 1995-07-18 | | Release date: | 1995-10-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a conserved protease that binds DNA: the bleomycin hydrolase, Gal6.
Science, 269, 1995
|
|
6UI4
 
 | | Crystal structure of phenamacril-bound F. graminearum myosin I | | Descriptor: | Calmodulin, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Zhou, Y, Zhou, X.E, Gong, Y, Zhu, Y, Xu, H.E, Zhou, M, Melcher, K, Zhang, F. | | Deposit date: | 2019-09-30 | | Release date: | 2020-03-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural basis of Fusarium myosin I inhibition by phenamacril.
Plos Pathog., 16, 2020
|
|
5JA5
 
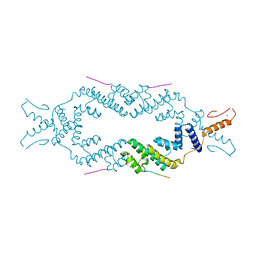 | | Crystal structure of the rice Topless related protein 2 (TPR2) N-terminal topless domain (1-209) L111A and L130A mutant in complex with rice D53 repressor EAR peptide motif | | Descriptor: | Protein TPR1, The rice D53 peptide (a.a. 794-808), ZINC ION | | Authors: | Ke, J, Ma, H, Gu, X, Brunzelle, J.S, Xu, H.E, Melcher, K. | | Deposit date: | 2016-04-12 | | Release date: | 2017-07-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A D53 repression motif induces oligomerization of TOPLESS corepressors and promotes assembly of a corepressor-nucleosome complex.
Sci Adv, 3, 2017
|
|
5JHP
 
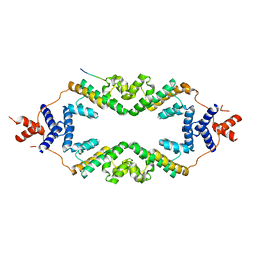 | | Crystal structure of the rice Topless related protein 2 (TPR2) N-terminal topless domain (1-209) L179A and I195A mutant in complex with rice D53 repressor EAR peptide motif | | Descriptor: | Protein TPR1, The rice D53 EAR peptide (794-808) | | Authors: | Ke, J, Ma, H, Gu, X, Brunzelle, J.S, Xu, H.E, Melcher, K. | | Deposit date: | 2016-04-21 | | Release date: | 2017-07-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | A D53 repression motif induces oligomerization of TOPLESS corepressors and promotes assembly of a corepressor-nucleosome complex.
Sci Adv, 3, 2017
|
|
5J9K
 
 | | Crystal structure of the rice Topless related protein 2 (TPR2) N-terminal topless domain (1-209) in complex with rice D53 repressor EAR peptide motif | | Descriptor: | Protein TPR1, ZINC ION, rice D53 peptide 794-808 | | Authors: | Ke, J, Ma, H, Gu, X, Brunzelle, J.S, Xu, H.E, Melcher, K. | | Deposit date: | 2016-04-10 | | Release date: | 2017-07-05 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | A D53 repression motif induces oligomerization of TOPLESS corepressors and promotes assembly of a corepressor-nucleosome complex.
Sci Adv, 3, 2017
|
|
8W89
 
 | | Cryo-EM structure of the PEA-bound TAAR1-Gs complex | | Descriptor: | 2-PHENYLETHYLAMINE, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Liu, H, Zheng, Y, Wang, Y, Wang, Y, He, X, Xu, P, Huang, S, Yuan, Q, Zhang, X, Wang, S, Xu, H.E, Xu, F. | | Deposit date: | 2023-09-01 | | Release date: | 2023-11-22 | | Last modified: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Recognition of methamphetamine and other amines by trace amine receptor TAAR1.
Nature, 624, 2023
|
|
