7XCZ
 
 | |
7XDK
 
 | | Cryo-EM structure of SARS-CoV-2 Delta Spike protein in complex with BA7054 and BA7125 fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BA7054 fab, ... | | Authors: | Liu, Z, Lui, S, Gao, Y. | | Deposit date: | 2022-03-27 | | Release date: | 2023-03-01 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Biparatopic antibody BA7208/7125 effectively neutralizes SARS-CoV-2 variants including Omicron BA.1-BA.5.
Cell Discov, 9, 2023
|
|
7XDA
 
 | |
7XDB
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron Spike protein in complex with BA7208 fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BA7208 fab, ... | | Authors: | Liu, Z, Liu, S, Gao, Y.Z. | | Deposit date: | 2022-03-26 | | Release date: | 2023-03-01 | | Method: | ELECTRON MICROSCOPY (2.62 Å) | | Cite: | Biparatopic antibody BA7208/7125 effectively neutralizes SARS-CoV-2 variants including Omicron BA.1-BA.5.
Cell Discov, 9, 2023
|
|
7XDL
 
 | | Cryo-EM structure of SARS-CoV-2 Delta Spike protein in complex with BA7208 and BA7125 fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BA7125 fab, ... | | Authors: | Liu, Z, Liu, S, Yuanzhu, G. | | Deposit date: | 2022-03-27 | | Release date: | 2023-03-15 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Biparatopic antibody BA7208/7125 effectively neutralizes SARS-CoV-2 variants including Omicron BA.1-BA.5.
Cell Discov, 9, 2023
|
|
8IUM
 
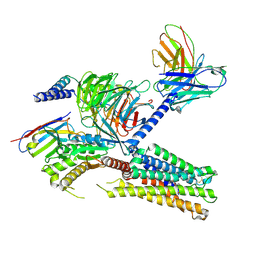 | | Cryo-EM structure of the tafluprost acid-bound human PTGFR-Gq complex | | Descriptor: | (~{Z})-7-[(1~{R},2~{R},3~{R},5~{S})-2-[(~{E})-3,3-bis(fluoranyl)-4-phenoxy-but-1-enyl]-3,5-bis(oxidanyl)cyclopentyl]hept-5-enoic acid, Antibody fragment scFv16, G subunit alpha (q), ... | | Authors: | Wu, C, Xu, Y, Xu, H.E. | | Deposit date: | 2023-03-24 | | Release date: | 2023-07-12 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | Ligand-induced activation and G protein coupling of prostaglandin F 2 alpha receptor.
Nat Commun, 14, 2023
|
|
8IUK
 
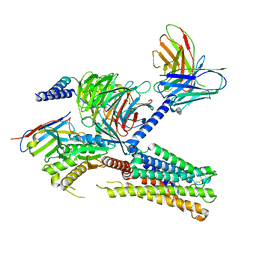 | | Cryo-EM structure of the PGF2-alpha-bound human PTGFR-Gq complex | | Descriptor: | (Z)-7-[(1R,2R,3R,5S)-3,5-bis(oxidanyl)-2-[(E,3S)-3-oxidanyloct-1-enyl]cyclopentyl]hept-5-enoic acid, Antibody fragment scFv16, G subunit alpha (q), ... | | Authors: | Wu, C, Xu, Y, Xu, H.E. | | Deposit date: | 2023-03-24 | | Release date: | 2023-07-12 | | Last modified: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (2.67 Å) | | Cite: | Ligand-induced activation and G protein coupling of prostaglandin F 2 alpha receptor.
Nat Commun, 14, 2023
|
|
8IUL
 
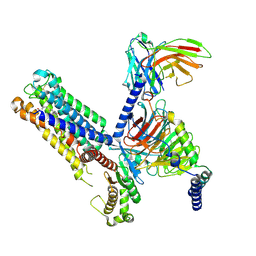 | | Cryo-EM structure of the latanoprost-bound human PTGFR-Gq complex | | Descriptor: | Antibody fragment scFv16, G subunit alpha (q), Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Wu, C, Xu, Y, Xu, H.E. | | Deposit date: | 2023-03-24 | | Release date: | 2023-07-12 | | Last modified: | 2023-08-30 | | Method: | ELECTRON MICROSCOPY (2.78 Å) | | Cite: | Ligand-induced activation and G protein coupling of prostaglandin F 2 alpha receptor.
Nat Commun, 14, 2023
|
|
3D5Q
 
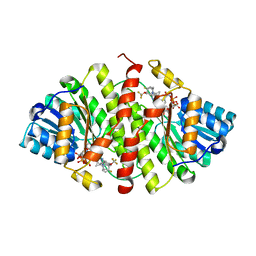 | | Crystal Structure of 11b-HSD1 in Complex with Triazole Inhibitor | | Descriptor: | 3-[1-(4-fluorophenyl)cyclopropyl]-4-(1-methylethyl)-5-[4-(trifluoromethoxy)phenyl]-4H-1,2,4-triazole, Corticosteroid 11-beta-dehydrogenase isozyme 1, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Wang, Z, Liu, J, Sudom, A, Walker, N.P.C. | | Deposit date: | 2008-05-16 | | Release date: | 2008-10-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Distinctive molecular inhibition mechanisms for selective inhibitors of human 11beta-hydroxysteroid dehydrogenase type 1.
Bioorg.Med.Chem., 16, 2008
|
|
8EWG
 
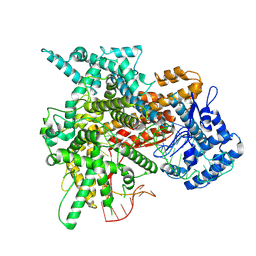 | | Cryo-EM structure of a riboendonclease | | Descriptor: | CRISPR-associated endonuclease Cas9, RNA (56-MER) | | Authors: | Li, Z, Wang, F. | | Deposit date: | 2022-10-23 | | Release date: | 2023-08-30 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural Basis for the Ribonuclease Activity of a Thermostable CRISPR-Cas13a from Thermoclostridium caenicola.
J.Mol.Biol., 435, 2023
|
|
2X6L
 
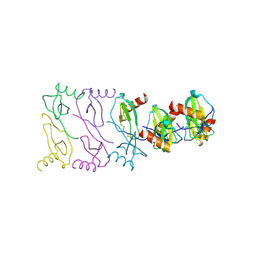 | |
2X69
 
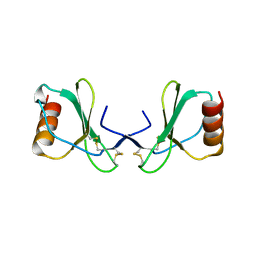 | |
2X6G
 
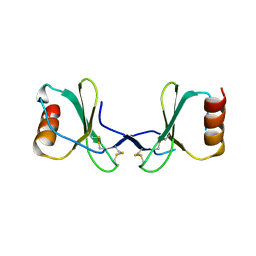 | |
8H8J
 
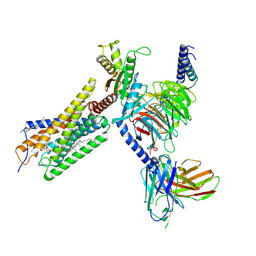 | | Lodoxamide-bound GPR35 in complex with G13 | | Descriptor: | 2,2'-[(2-chloro-5-cyano-1,3-phenylene)bis(azanediyl)]bis(oxoacetic acid), CALCIUM ION, CHOLESTEROL, ... | | Authors: | Yuan, Q, Duan, J, Liu, Q, Xu, H.E, Jiang, Y. | | Deposit date: | 2022-10-23 | | Release date: | 2023-02-08 | | Last modified: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Insights into divalent cation regulation and G 13 -coupling of orphan receptor GPR35.
Cell Discov, 8, 2022
|
|
4M6T
 
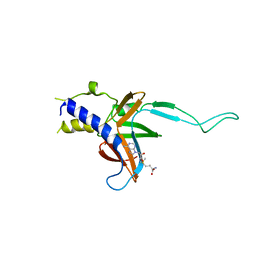 | | Structure of human Paf1 and Leo1 complex | | Descriptor: | RNA polymerase II-associated factor 1 homolog, Linker, RNA polymerase-associated protein LEO1, ... | | Authors: | Shen, Y, Qin, X. | | Deposit date: | 2013-08-11 | | Release date: | 2013-10-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.498 Å) | | Cite: | Structural insights into Paf1 complex assembly and histone binding
Nucleic Acids Res., 41, 2013
|
|
3QJ8
 
 | |
3QKV
 
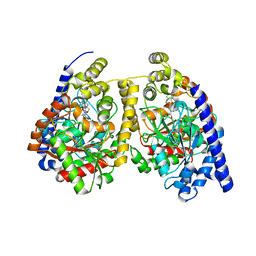 | | Crystal structure of fatty acid amide hydrolase with small molecule compound | | Descriptor: | (6-bromo-1'H,4H-spiro[1,3-benzodioxine-2,4'-piperidin]-1'-yl)methanol, Fatty-acid amide hydrolase 1 | | Authors: | Min, X, Walker, N.P.C, Wang, Z. | | Deposit date: | 2011-02-01 | | Release date: | 2011-04-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Discovery and molecular basis of potent noncovalent inhibitors of fatty acid amide hydrolase (FAAH).
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3QJ9
 
 | | Crystal structure of fatty acid amide hydrolase with small molecule inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 1-{(3S)-1-[4-(1-benzofuran-2-yl)pyrimidin-2-yl]piperidin-3-yl}-3-ethyl-1,3-dihydro-2H-benzimidazol-2-one, Fatty-acid amide hydrolase 1, ... | | Authors: | Min, X, Walker, N.P.C, Wang, Z. | | Deposit date: | 2011-01-28 | | Release date: | 2011-04-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery and molecular basis of potent noncovalent inhibitors of fatty acid amide hydrolase (FAAH).
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
8XZG
 
 | | Cryo-EM structure of the [Pyr1]-apelin-13-bound human APLNR-Gi complex | | Descriptor: | Apelin receptor, Apelin-13, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Wang, W, Ji, S, Zhang, Y. | | Deposit date: | 2024-01-21 | | Release date: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure-based design of non-hypertrophic apelin receptor modulator.
Cell, 187, 2024
|
|
8XZF
 
 | | Cryo-EM structure of the WN561-bound human APLNR-Gi complex | | Descriptor: | Apelin receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Wang, W, Ji, S, Zhang, Y. | | Deposit date: | 2024-01-21 | | Release date: | 2024-03-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure-based design of non-hypertrophic apelin receptor modulator.
Cell, 187, 2024
|
|
7XLQ
 
 | | Structure of human R-type voltage-gated CaV2.3-alpha2/delta1-beta1 channel complex in the ligand-free (apo) state | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Gao, Y, Qiu, Y, Wei, Y, Dong, Y, Zhang, X.C, Zhao, Y. | | Deposit date: | 2022-04-22 | | Release date: | 2023-02-08 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Molecular insights into the gating mechanisms of voltage-gated calcium channel Ca V 2.3.
Nat Commun, 14, 2023
|
|
8XZJ
 
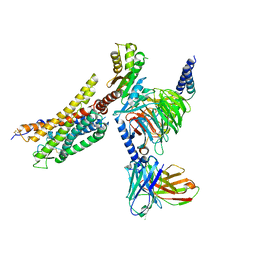 | | Cryo-EM structure of the WN353-bound human APLNR-Gi complex | | Descriptor: | Apelin receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Wang, W, Ji, S, Zhang, Y. | | Deposit date: | 2024-01-21 | | Release date: | 2024-03-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure-based design of non-hypertrophic apelin receptor modulator.
Cell, 187, 2024
|
|
8XZI
 
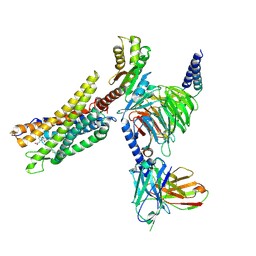 | | Cryo-EM structure of the CMF-019-bound human APLNR-Gi complex | | Descriptor: | (3~{S})-5-methyl-3-[[1-pentan-3-yl-2-(thiophen-2-ylmethyl)benzimidazol-5-yl]carbonylamino]hexanoic acid, Apelin receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Wang, W, Ji, S, Zhang, Y. | | Deposit date: | 2024-01-21 | | Release date: | 2024-03-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structure-based design of non-hypertrophic apelin receptor modulator.
Cell, 187, 2024
|
|
8XZH
 
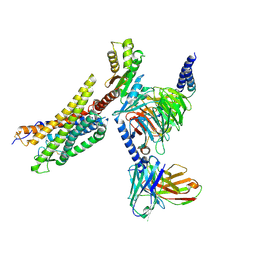 | | Cryo-EM structure of the MM07-bound human APLNR-Gi complex | | Descriptor: | Apelin receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Wang, W, Ji, S, Zhang, Y. | | Deposit date: | 2024-01-21 | | Release date: | 2024-03-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structure-based design of non-hypertrophic apelin receptor modulator.
Cell, 187, 2024
|
|
6USF
 
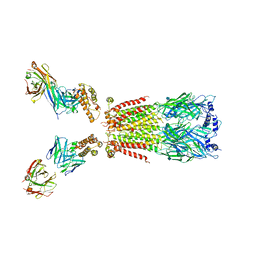 | | CryoEM structure of human alpha4beta2 nicotinic acetylcholine receptor with varenicline in complex with anti-BRIL synthetic antibody BAK5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab heavy chain, ... | | Authors: | Alvarez, F.J.D, Mukherjee, S, Han, S, Ammirati, M, Kossiakoff, A.A. | | Deposit date: | 2019-10-26 | | Release date: | 2020-04-29 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.87 Å) | | Cite: | Synthetic antibodies against BRIL as universal fiducial marks for single-particle cryoEM structure determination of membrane proteins.
Nat Commun, 11, 2020
|
|
