6W4H
 
 | | 1.80 Angstrom Resolution Crystal Structure of NSP16 - NSP10 Complex from SARS-CoV-2 | | Descriptor: | 2'-O-methyltransferase, ACETATE ION, Non-structural protein 10, ... | | Authors: | Minasov, G, Shuvalova, L, Rosas-Lemus, M, Kiryukhina, O, Wiersum, G, Godzik, A, Jaroszewski, L, Stogios, P.J, Skarina, T, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-03-10 | | Release date: | 2020-03-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | High-resolution structures of the SARS-CoV-2 2'- O -methyltransferase reveal strategies for structure-based inhibitor design.
Sci.Signal., 13, 2020
|
|
6W75
 
 | | 1.95 Angstrom Resolution Crystal Structure of NSP10 - NSP16 Complex from SARS-CoV-2 | | Descriptor: | 2'-O-methyltransferase, FORMIC ACID, Non-structural protein 10, ... | | Authors: | Minasov, G, Shuvalova, L, Rosas-Lemus, M, Kiryukhina, O, Wiersum, G, Godzik, A, Jaroszewski, L, Stogios, P.J, Skarina, T, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-03-18 | | Release date: | 2020-03-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.951 Å) | | Cite: | High-resolution structures of the SARS-CoV-2 2'- O -methyltransferase reveal strategies for structure-based inhibitor design.
Sci.Signal., 13, 2020
|
|
6WQ3
 
 | | Crystal Structure of Nsp16-Nsp10 Heterodimer from SARS-CoV-2 in Complex with 7-methyl-GpppA and S-adenosyl-L-homocysteine. | | Descriptor: | 2'-O-methyltransferase, 7N-METHYL-8-HYDROGUANOSINE-5'-DIPHOSPHATE, Non-structural protein 10, ... | | Authors: | Minasov, G, Shuvalova, L, Rosas-Lemus, M, Brunzelle, J.S, Kiryukhina, O, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-04-28 | | Release date: | 2020-05-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | High-resolution structures of the SARS-CoV-2 2'- O -methyltransferase reveal strategies for structure-based inhibitor design.
Sci.Signal., 13, 2020
|
|
6WRZ
 
 | | Crystal Structure of Nsp16-Nsp10 Heterodimer from SARS-CoV-2 with 7-methyl-GpppA and S-adenosyl-L-homocysteine in the Active Site and Sulfates in the mRNA Binding Groove. | | Descriptor: | 2'-O-methyltransferase, 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Minasov, G, Shuvalova, L, Rosas-Lemus, M, Brunzelle, J.S, Kiryukhina, O, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-04-30 | | Release date: | 2020-05-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | High-resolution structures of the SARS-CoV-2 2'- O -methyltransferase reveal strategies for structure-based inhibitor design.
Sci.Signal., 13, 2020
|
|
7JZ0
 
 | | Crystal Structure of SARS-CoV-2 Nsp16/10 Heterodimer in Complex with (m7GpppA2m)pUpUpApApA (Cap-1) and S-Adenosyl-L-homocysteine (SAH). | | Descriptor: | 2'-O-methyltransferase, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Minasov, G, Shuvalova, L, Rosas-Lemus, M, Kiryukhina, O, Brunzelle, J.S, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-09-01 | | Release date: | 2020-09-16 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure of SARS-CoV-2 2'-O-methyltransferase heterodimer with RNA Cap analog and sulfates bound reveals new strategies for structure-based inhibitor design
Biorxiv, 2020
|
|
6WJT
 
 | | 2.0 Angstrom Resolution Crystal Structure of Nsp16-Nsp10 Heterodimer from SARS-CoV-2 in Complex with S-Adenosyl-L-Homocysteine | | Descriptor: | 2'-O-methyltransferase, FORMIC ACID, Non-structural protein 10, ... | | Authors: | Minasov, G, Shuvalova, L, Rosas-Lemus, M, Kiryukhina, O, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-04-14 | | Release date: | 2020-04-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | High-resolution structures of the SARS-CoV-2 2'- O -methyltransferase reveal strategies for structure-based inhibitor design.
Sci.Signal., 13, 2020
|
|
6WKQ
 
 | | 1.98 Angstrom Resolution Crystal Structure of NSP16-NSP10 Heterodimer from SARS-CoV-2 in Complex with Sinefungin | | Descriptor: | 2'-O-methyltransferase, FORMIC ACID, Non-structural protein 10, ... | | Authors: | Minasov, G, Shuvalova, L, Rosas-Lemus, M, Kiryukhina, O, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-04-16 | | Release date: | 2020-04-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | High-resolution structures of the SARS-CoV-2 2'- O -methyltransferase reveal strategies for structure-based inhibitor design.
Sci.Signal., 13, 2020
|
|
6WVN
 
 | | Crystal Structure of Nsp16-Nsp10 from SARS-CoV-2 in Complex with 7-methyl-GpppA and S-Adenosylmethionine. | | Descriptor: | 2'-O-methyltransferase, 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE, ADENINE, ... | | Authors: | Minasov, G, Shuvalova, L, Rosas-Lemus, M, Kiryukhina, O, Brunzelle, J.S, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-05-06 | | Release date: | 2020-05-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | High-resolution structures of the SARS-CoV-2 2'- O -methyltransferase reveal strategies for structure-based inhibitor design.
Sci.Signal., 13, 2020
|
|
7N1M
 
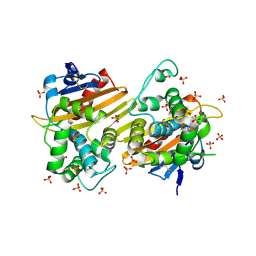 | | Crystal Structure of the Class D Beta-lactamase OXA-935 from Pseudomonas aeruginosa, Orthorhombic Crystal Form | | Descriptor: | Beta-lactamase OXA-935, SULFATE ION | | Authors: | Minasov, G, Shuvalova, L, Rosas-Lemus, M, Brunzelle, J.B, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-05-27 | | Release date: | 2022-07-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Functional and Structural Characterization of OXA-935, a Novel OXA-10-Family beta-Lactamase from Pseudomonas aeruginosa.
Antimicrob.Agents Chemother., 66, 2022
|
|
7RL8
 
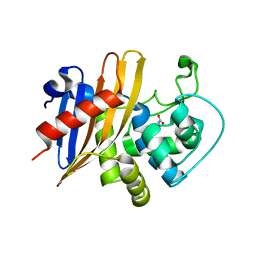 | | Crystal Structure of C79A Mutant of Class D beta-lactamase from Clostridium difficile 630 | | Descriptor: | Beta-lactamase, DI(HYDROXYETHYL)ETHER, SULFATE ION | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Rosas-Lemus, M, Jedrzejczak, R, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-07-23 | | Release date: | 2021-08-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of C79A Mutant of Class D beta-lactamase from Clostridium difficile 630
To Be Published
|
|
7RLR
 
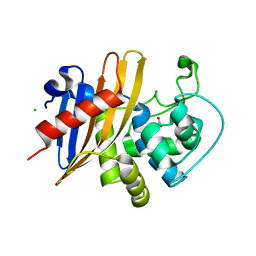 | | Crystal Structure of K83A Mutant of Class D beta-lactamase from Clostridium difficile 630 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Beta-lactamase, ... | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Rosas-Lemus, M, Jedrzejczak, R, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-07-26 | | Release date: | 2021-08-11 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Crystal Structure of K83A Mutant of Class D beta-lactamase from Clostridium difficile 630
To Be Published
|
|
8F4S
 
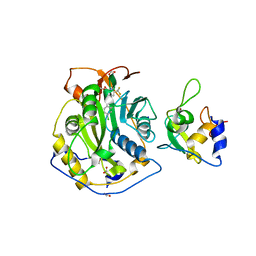 | | Crystal Structure of the SARS-CoV-2 2'-O-Methyltransferase with Compound 5a bound to the Cryptic Pocket of nsp16 | | Descriptor: | 2'-O-methyltransferase, 4-[(E)-2-(2,4-dichlorophenyl)ethenyl]-6-(trifluoromethyl)pyrimidin-2-ol, FORMIC ACID, ... | | Authors: | Minasov, G, Shuvalova, L, Brunzelle, J.S, Rosas-Lemus, M, Kiryukhina, O, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2022-11-11 | | Release date: | 2023-10-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Discovery of a Druggable, Cryptic Pocket in SARS-CoV-2 nsp16 Using Allosteric Inhibitors.
Acs Infect Dis., 9, 2023
|
|
8F4Y
 
 | | Crystal Structure of SARS-CoV-2 2'-O-Methyltransferase in Complex with Compound 5a covalently bound to nsp16 and nsp10 | | Descriptor: | 2'-O-methyltransferase, 4-[(E)-2-(2,4-dichlorophenyl)ethenyl]-6-(trifluoromethyl)pyrimidin-2-ol, 4-[2-(2,4-dichlorophenyl)ethyl]-6-(trifluoromethyl)pyrimidin-2-ol, ... | | Authors: | Minasov, G, Shuvalova, L, Brunzelle, J.S, Rosas-Lemus, M, Kiryukhina, O, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2022-11-11 | | Release date: | 2023-10-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Discovery of a Druggable, Cryptic Pocket in SARS-CoV-2 nsp16 Using Allosteric Inhibitors.
Acs Infect Dis., 9, 2023
|
|
7ULT
 
 | | Crystal Structure of SARS-CoV-2 Nsp16/10 Heterodimer Apo-Form. | | Descriptor: | 2'-O-methyltransferase, FORMIC ACID, Non-structural protein 10, ... | | Authors: | Minasov, G, Shuvalova, L, Brunzelle, J.S, Rosas-Lemus, M, Kiryukhina, O, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-04-05 | | Release date: | 2022-04-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of SARS-CoV-2 Nsp16/10 Heterodimer Apo-Form.
To Be Published
|
|
8UFM
 
 | | Crystal Structure of L516C/Y647C Mutant of SARS-Unique Domain (SUD) from SARS-CoV-2 | | Descriptor: | ACETATE ION, FORMIC ACID, Papain-like protease nsp3, ... | | Authors: | Minasov, G, Shuvalova, L, Brunzelle, J.S, Rosas-Lemus, M, Kiryukhina, O, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID), Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2023-10-04 | | Release date: | 2023-10-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal Structure of L516C/Y647C Mutant of SARS-Unique Domain (SUD) from SARS-CoV-2
To Be Published
|
|
8UFL
 
 | | Crystal Structure of SARS-Unique Domain (SUD) of Nsp3 from SARS coronavirus | | Descriptor: | CHLORIDE ION, Papain-like protease nsp3, SULFATE ION | | Authors: | Minasov, G, Shuvalova, L, Rosas-Lemus, M, Kiryukhina, O, Brunzelle, J.S, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID), Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2023-10-04 | | Release date: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Crystal Structure of SARS-Unique Domain (SUD) of Nsp3 from SARS coronavirus
To Be Published
|
|
6XKM
 
 | | Room Temperature Structure of SARS-CoV-2 NSP10/NSP16 Methyltransferase in a Complex with SAM Determined by Fixed-Target Serial Crystallography | | Descriptor: | 2'-O-methyltransferase, CHLORIDE ION, Non-structural protein 10, ... | | Authors: | Wilamowski, M, Sherrell, D.A, Minasov, G, Kim, Y, Shuvalova, L, Lavens, A, Chard, R, Rosas-Lemus, M, Maltseva, N, Jedrzejczak, R, Michalska, K, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-06-26 | | Release date: | 2020-07-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | 2'-O methylation of RNA cap in SARS-CoV-2 captured by serial crystallography.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
8SFG
 
 | | Crystal Structure of the Open Unbound Catalytically Inactive Makes Caterpillars Floppy-like (MCF) Effector from Vibrio vulnificus CMCP6 | | Descriptor: | Autotransporter adhesin, CHLORIDE ION, SULFATE ION | | Authors: | Minasov, G, Shuvalova, L, Rosas-Lemus, M, Herrera, A, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2023-04-11 | | Release date: | 2024-06-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of the Open Unbound Catalytically Inactive Makes Caterpillars Floppy-like (MCF) Effector from Vibrio vulnificus CMCP6.
To Be Published
|
|
6UE2
 
 | | 1.85 Angstrom Resolution Crystal Structure of Class D beta-lactamase from Clostridium difficile 630 | | Descriptor: | Beta-lactamase, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Rosas-Lemus, M, Jedrzejczak, R, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-09-20 | | Release date: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | 1.85 Angstrom Resolution Crystal Structure of Class D beta-lactamase from Clostridium difficile 630.
To Be Published
|
|
6WY4
 
 | | Crystal Structure of Wild Type Class D beta-lactamase from Clostridium difficile 630 | | Descriptor: | Beta-lactamase, DI(HYDROXYETHYL)ETHER, SODIUM ION | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Rosas-Lemus, M, Jedrzejczak, R, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-05-12 | | Release date: | 2020-05-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Wild Type Class D beta-lactamase from Clostridium difficile 630
To Be Published
|
|
7KOA
 
 | | Room Temperature Structure of SARS-CoV-2 Nsp10/16 Methyltransferase in a Complex with Cap-0 and SAM Determined by Pink-Beam Serial Crystallography | | Descriptor: | 2'-O-methyltransferase, Non-structural protein 10, P1-7-METHYLGUANOSINE-P3-ADENOSINE-5',5'-TRIPHOSPHATE, ... | | Authors: | Wilamowski, M, Sherrell, D.A, Minasov, G, Shuvalova, L, Lavens, A, Henning, R, Maltseva, N, Rosas-Lemus, M, Kim, Y, Satchell, K.J.F, Srajer, V, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-11-07 | | Release date: | 2020-11-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Room Temperature Structure of SARS-CoV-2 Nsp10/Nsp16 Methyltransferase in a Complex with Cap-0 and SAM Determined by Pink-Beam Serial Crystallography
To Be Published
|
|
7JPE
 
 | | Room Temperature Structure of SARS-CoV-2 Nsp10/Nsp16 Methyltransferase in a Complex with m7GpppA Cap-0 and SAM Determined by Fixed-Target Serial Crystallography | | Descriptor: | 2'-O-methyltransferase, 7N-METHYL-8-HYDROGUANOSINE-5'-DIPHOSPHATE, Non-structural protein 10, ... | | Authors: | Wilamowski, M, Sherrell, D.A, Minasov, G, Kim, Y, Shuvalova, L, Lavens, A, Chard, R, Rosas-Lemus, M, Maltseva, N, Jedrzejczak, R, Michalska, K, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-08-07 | | Release date: | 2020-08-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | 2'-O methylation of RNA cap in SARS-CoV-2 captured by serial crystallography.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7JIB
 
 | | Room Temperature Crystal Structure of Nsp10/Nsp16 from SARS-CoV-2 with Substrates and Products of 2'-O-methylation of the Cap-1 | | Descriptor: | 2'-O-methyltransferase, 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE, 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE-5'-(2'-O-METHYL)-ADENOSINE, ... | | Authors: | Wilamowski, M, Minasov, G, Kim, Y, Sherrell, D.A, Shuvalova, L, Lavens, A, Chard, R, Rosas-Lemus, M, Maltseva, N, Jedrzejczak, R, Michalska, K, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-07-23 | | Release date: | 2020-08-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | 2'-O methylation of RNA cap in SARS-CoV-2 captured by serial crystallography.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7JHE
 
 | | Room Temperature Structure of SARS-CoV-2 Nsp10/Nsp16 Methyltransferase in a Complex with 2'-O-methylated m7GpppA Cap-1 and SAH Determined by Fixed-Target Serial Crystallography | | Descriptor: | 2'-O-methyltransferase, 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE, 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE-5'-(2'-O-METHYL)-ADENOSINE, ... | | Authors: | Wilamowski, M, Sherrell, D.A, Minasov, G, Kim, Y, Shuvalova, L, Lavens, A, Chard, R, Rosas-Lemus, M, Maltseva, N, Jedrzejczak, R, Michalska, K, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-07-20 | | Release date: | 2020-08-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | 2'-O methylation of RNA cap in SARS-CoV-2 captured by serial crystallography.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7L5R
 
 | | Crystal Structure of the Oxacillin-hydrolyzing Class D Extended-spectrum Beta-lactamase OXA-14 from Pseudomonas aeruginosa | | Descriptor: | Beta-lactamase, GLYCEROL, SULFATE ION | | Authors: | Minasov, G, Shuvalova, L, Rosas-Lemus, M, Brunzelle, J.S, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-12-22 | | Release date: | 2021-12-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Functional and Structural Characterization of OXA-935, a Novel OXA-10-Family beta-Lactamase from Pseudomonas aeruginosa.
Antimicrob.Agents Chemother., 66, 2022
|
|
