1F2N
 
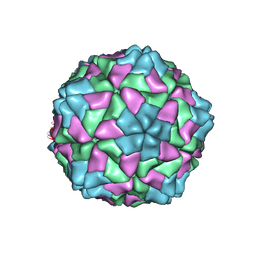 | | RICE YELLOW MOTTLE VIRUS | | Descriptor: | CALCIUM ION, CAPSID PROTEIN | | Authors: | Qu, C, Liljas, L, Opalka, N, Brugidou, C, Yeager, M, Beachy, R.N, Fauquet, C.M, Johnson, J.E, Lin, T. | | Deposit date: | 2000-05-26 | | Release date: | 2000-11-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | 3D domain swapping modulates the stability of members of an icosahedral virus group.
Structure Fold.Des., 8, 2000
|
|
1OSI
 
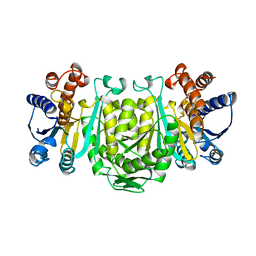 | | STRUCTURE OF 3-ISOPROPYLMALATE DEHYDROGENASE | | Descriptor: | 3-ISOPROPYLMALATE DEHYDROGENASE | | Authors: | Qu, C, Akanuma, S, Moriyama, H, Tanaka, N, Oshima, T. | | Deposit date: | 1996-10-22 | | Release date: | 1997-01-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A mutation at the interface between domains causes rearrangement of domains in 3-isopropylmalate dehydrogenase.
Protein Eng., 10, 1997
|
|
1OSJ
 
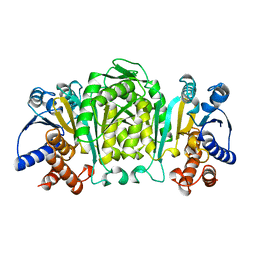 | | STRUCTURE OF 3-ISOPROPYLMALATE DEHYDROGENASE | | Descriptor: | 3-ISOPROPYLMALATE DEHYDROGENASE | | Authors: | Qu, C, Akanuma, S, Moriyama, H, Tanaka, N, Oshima, T. | | Deposit date: | 1996-10-22 | | Release date: | 1997-01-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | A mutation at the interface between domains causes rearrangement of domains in 3-isopropylmalate dehydrogenase.
Protein Eng., 10, 1997
|
|
1G2U
 
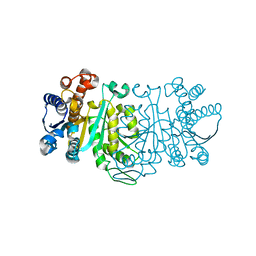 | | THE STRUCTURE OF THE MUTANT, A172V, OF 3-ISOPROPYLMALATE DEHYDROGENASE FROM THERMUS THERMOPHILUS HB8 : ITS THERMOSTABILITY AND STRUCTURE. | | Descriptor: | 3-ISOPROPYLMALATE DEHYDROGENASE | | Authors: | Qu, C, Akanuma, S, Tanaka, N, Moriyama, H, Oshima, T. | | Deposit date: | 2000-10-21 | | Release date: | 2000-11-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Design, X-ray crystallography, molecular modelling and thermal stability studies of mutant enzymes at site 172 of 3-isopropylmalate dehydrogenase from Thermus thermophilus.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1GC8
 
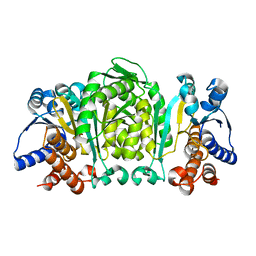 | | THE CRYSTAL STRUCTURE OF THERMUS THERMOPHILUS 3-ISOPROPYLMALATE DEHYDROGENASE MUTATED AT 172TH FROM ALA TO PHE | | Descriptor: | 3-ISOPROPYLMALATE DEHYDROGENASE | | Authors: | Qu, C, Akanuma, S, Tanaka, N, Moriyama, H, Oshima, T. | | Deposit date: | 2000-07-27 | | Release date: | 2000-09-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Design, X-ray crystallography, molecular modelling and thermal stability studies of mutant enzymes at site 172 of 3-isopropylmalate dehydrogenase from Thermus thermophilus.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1GC9
 
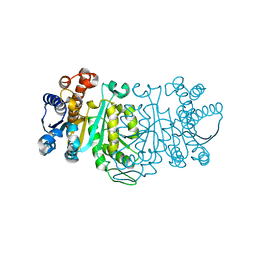 | | THE CRYSTAL STRUCTURE OF THERMUS THERMOPHILUS 3-ISOPROPYLMALATE DEHYDROGENASE MUTATED AT 172TH FROM ALA TO GLY | | Descriptor: | 3-ISOPROPYLMALATE DEHYDROGENASE | | Authors: | Qu, C, Akanuma, S, Tanaka, N, Moriyama, H, Oshima, T. | | Deposit date: | 2000-07-28 | | Release date: | 2000-09-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Design, X-ray crystallography, molecular modelling and thermal stability studies of mutant enzymes at site 172 of 3-isopropylmalate dehydrogenase from Thermus thermophilus.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
7Y55
 
 | |
7CX4
 
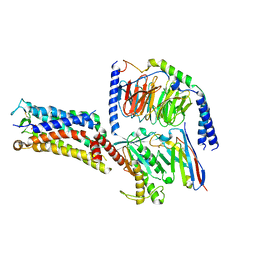 | | Cryo-EM structure of the Evatanepag-bound EP2-Gs complex | | Descriptor: | 2-[3-[[(4-~{tert}-butylphenyl)methyl-pyridin-3-ylsulfonyl-amino]methyl]phenoxy]ethanoic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Qu, C, Mao, C, Xiao, P, Shen, Q, Zhong, Y, Yang, F, Shen, D, Tao, X, Zhang, H, Yan, X, Zhao, R, He, J, Guan, Y, Zhang, C, Hou, G, Zhang, P, Yu, X, Guan, Y, Sun, J, Zhang, Y. | | Deposit date: | 2020-09-01 | | Release date: | 2021-05-05 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Ligand recognition, unconventional activation, and G protein coupling of the prostaglandin E 2 receptor EP2 subtype.
Sci Adv, 7, 2021
|
|
7CX3
 
 | | Cryo-EM structure of the Taprenepag-bound EP2-Gs complex | | Descriptor: | 2-[3-[[(4-pyrazol-1-ylphenyl)methyl-pyridin-3-ylsulfonyl-amino]methyl]phenoxy]ethanoic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Qu, C, Mao, C, Xiao, P, Shen, Q, Zhong, Y, Yang, F, Shen, D, Tao, X, Zhang, H, Yan, X, Zhao, R, He, J, Guan, Y, Zhang, C, Hou, G, Zhang, P, Yu, X, Guan, Y, Sun, J, Zhang, Y. | | Deposit date: | 2020-09-01 | | Release date: | 2021-05-05 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Ligand recognition, unconventional activation, and G protein coupling of the prostaglandin E 2 receptor EP2 subtype.
Sci Adv, 7, 2021
|
|
7CX2
 
 | | Cryo-EM structure of the PGE2-bound EP2-Gs complex | | Descriptor: | (Z)-7-[(1R,2R,3R)-3-hydroxy-2-[(E,3S)-3-hydroxyoct-1-enyl]-5-oxo-cyclopentyl]hept-5-enoic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Qu, C, Mao, C, Xiao, P, Shen, Q, Zhong, Y, Yang, F, Shen, D, Tao, X, Zhang, H, Yan, X, Zhao, R, He, J, Guan, Y, Zhang, C, Hou, G, Zhang, P, Yu, X, Guan, Y, Sun, J, Zhang, Y. | | Deposit date: | 2020-09-01 | | Release date: | 2021-05-05 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Ligand recognition, unconventional activation, and G protein coupling of the prostaglandin E 2 receptor EP2 subtype.
Sci Adv, 7, 2021
|
|
1ZA7
 
 | | The crystal structure of salt stable cowpea cholorotic mottle virus at 2.7 angstroms resolution. | | Descriptor: | Coat protein | | Authors: | Bothner, B, Speir, J.A, Qu, C, Willits, D.A, Young, M.J, Johnson, J.E. | | Deposit date: | 2005-04-05 | | Release date: | 2006-03-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Enhanced local symmetry interactions globally stabilize a mutant virus capsid that maintains infectivity and capsid dynamics.
J.Virol., 80, 2006
|
|
7D77
 
 | | Cryo-EM structure of the cortisol-bound adhesion receptor GPR97-Go complex | | Descriptor: | (11alpha,14beta)-11,17,21-trihydroxypregn-4-ene-3,20-dione, Adhesion G protein-coupled receptor G3; GPR97, CHOLESTEROL, ... | | Authors: | Ping, Y, Mao, C, Xiao, P, Zhao, R, Jiang, Y, Yang, Z, An, W, Shen, D, Yang, F, Zhang, H, Qu, C, Shen, Q, Tian, C, Li, Z, Li, S, Wang, G, Tao, X, Wen, X, Zhong, Y, Yang, J, Yi, F, Yu, X, Xu, E, Zhang, Y, Sun, J. | | Deposit date: | 2020-10-03 | | Release date: | 2021-02-03 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structures of the glucocorticoid-bound adhesion receptor GPR97-G o complex.
Nature, 589, 2021
|
|
7D76
 
 | | Cryo-EM structure of the beclomethasone-bound adhesion receptor GPR97-Go complex | | Descriptor: | (8~{S},9~{R},10~{S},11~{S},13~{S},14~{S},16~{S},17~{R})-9-chloranyl-10,13,16-trimethyl-11,17-bis(oxidanyl)-17-(2-oxidanylethanoyl)-6,7,8,11,12,14,15,16-octahydrocyclopenta[a]phenanthren-3-one, Adhesion G protein-coupled receptor G3; GPR97, CHOLESTEROL, ... | | Authors: | Ping, Y, Mao, C, Xiao, P, Zhao, R, Jiang, Y, Yang, Z, An, W, Shen, D, Yang, F, Zhang, H, Qu, C, Shen, Q, Tian, C, Li, Z, Li, S, Wang, G, Tao, X, Wen, X, Zhong, Y, Yang, J, Yi, F, Yu, X, Xu, E, Zhang, Y, Sun, J. | | Deposit date: | 2020-10-03 | | Release date: | 2021-02-03 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structures of the glucocorticoid-bound adhesion receptor GPR97-G o complex.
Nature, 589, 2021
|
|
5IKK
 
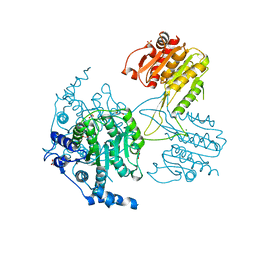 | | Structure of the histone deacetylase Clr3 | | Descriptor: | 1,2-ETHANEDIOL, Histone deacetylase clr3, MAGNESIUM ION, ... | | Authors: | Brugger, C, Schalch, T. | | Deposit date: | 2016-03-03 | | Release date: | 2016-04-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | SHREC Silences Heterochromatin via Distinct Remodeling and Deacetylation Modules.
Mol.Cell, 62, 2016
|
|
5IKF
 
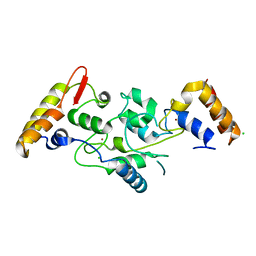 | |
6WTQ
 
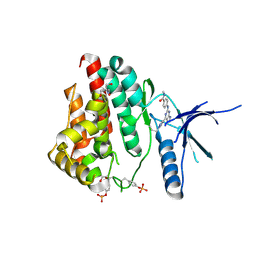 | | Human JAK2 JH1 domain in complex with PROTAC-intermediate linker handle 4 | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, N-methyl-4-{[4-(1-propyl-1H-pyrazol-4-yl)-7H-pyrrolo[2,3-d]pyrimidin-2-yl]amino}benzamide, ... | | Authors: | Yu, S, Nithianantham, S, Fischer, M. | | Deposit date: | 2020-05-03 | | Release date: | 2021-05-05 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.79968476 Å) | | Cite: | Degradation of Janus kinases in CRLF2-rearranged acute lymphoblastic leukemia.
Blood, 138, 2021
|
|
6WTN
 
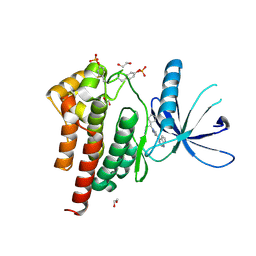 | | Human JAK2 JH1 domain in complex with Ruxolitinib | | Descriptor: | (3R)-3-cyclopentyl-3-[4-(7H-pyrrolo[2,3-d]pyrimidin-4-yl)-1H-pyrazol-1-yl]propanenitrile, 1,2-ETHANEDIOL, Tyrosine-protein kinase JAK2 | | Authors: | Yu, S, Nithianantham, S, Fischer, M. | | Deposit date: | 2020-05-03 | | Release date: | 2021-05-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Degradation of Janus kinases in CRLF2-rearranged acute lymphoblastic leukemia.
Blood, 138, 2021
|
|
6WTP
 
 | | Human JAK2 JH1 domain in complex with PROTAC-intermediate linker handle 3 | | Descriptor: | GLYCEROL, Tyrosine-protein kinase JAK2, tert-butyl 4-[(4-{1-[3-(cyanomethyl)-1-(ethylsulfonyl)azetidin-3-yl]-1H-pyrazol-4-yl}-7H-pyrrolo[2,3-d]pyrimidin-2-yl)amino]benzoate | | Authors: | Yu, S, Nithianantham, S, Fischer, M. | | Deposit date: | 2020-05-03 | | Release date: | 2021-05-05 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Degradation of Janus kinases in CRLF2-rearranged acute lymphoblastic leukemia.
Blood, 138, 2021
|
|
6WTO
 
 | | Human JAK2 JH1 domain in complex with Baricitinib | | Descriptor: | 1,2-ETHANEDIOL, Baricitinib, Tyrosine-protein kinase JAK2 | | Authors: | Yu, S, Nithianantham, S, Fischer, M. | | Deposit date: | 2020-05-03 | | Release date: | 2021-05-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Degradation of Janus kinases in CRLF2-rearranged acute lymphoblastic leukemia.
Blood, 138, 2021
|
|
5IKJ
 
 | | Structure of Clr2 bound to the Clr1 C-terminus | | Descriptor: | CHLORIDE ION, Cryptic loci regulator 2, Cryptic loci regulator protein 1, ... | | Authors: | Pfister, Y, Schalch, T. | | Deposit date: | 2016-03-03 | | Release date: | 2016-04-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | SHREC Silences Heterochromatin via Distinct Remodeling and Deacetylation Modules.
Mol.Cell, 62, 2016
|
|
7CRH
 
 | | Cryo-EM structure of SKF83959 bound dopamine receptor DRD1-Gs signaling complex | | Descriptor: | (1S)-6-chloranyl-3-methyl-1-(3-methylphenyl)-1,2,4,5-tetrahydro-3-benzazepine-7,8-diol, D(1A) dopamine receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Yan, W, Shao, Z.H. | | Deposit date: | 2020-08-13 | | Release date: | 2021-03-03 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Ligand recognition and allosteric regulation of DRD1-Gs signaling complexes.
Cell, 184, 2021
|
|
7CKW
 
 | | Cryo-EM structure of Fenoldopam bound dopamine receptor DRD1-Gs signaling complex | | Descriptor: | (1R)-6-chloranyl-1-(4-hydroxyphenyl)-2,3,4,5-tetrahydro-1H-3-benzazepine-7,8-diol, CHOLESTEROL, D(1A) dopamine receptor, ... | | Authors: | Yan, W, Shao, W. | | Deposit date: | 2020-07-20 | | Release date: | 2021-03-03 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Ligand recognition and allosteric regulation of DRD1-Gs signaling complexes.
Cell, 184, 2021
|
|
7CKY
 
 | | Cryo-EM structure of PW0464 bound dopamine receptor DRD1-Gs signaling complex | | Descriptor: | 6-[4-[3-[bis(fluoranyl)methoxy]pyridin-2-yl]oxy-2-methyl-phenyl]-1,5-dimethyl-pyrimidine-2,4-dione, CHOLESTEROL, D(1A) dopamine receptor, ... | | Authors: | Yan, W, Shao, Z. | | Deposit date: | 2020-07-20 | | Release date: | 2021-03-03 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Ligand recognition and allosteric regulation of DRD1-Gs signaling complexes.
Cell, 184, 2021
|
|
7CKZ
 
 | | Cryo-EM structure of Dopamine and LY3154207 bound dopamine receptor DRD1-Gs signaling complex | | Descriptor: | 2-[2,6-bis(chloranyl)phenyl]-1-[(1S,3R)-3-(hydroxymethyl)-1-methyl-5-(3-methyl-3-oxidanyl-butyl)-3,4-dihydro-1H-isoquinolin-2-yl]ethanone, CHOLESTEROL, D(1A) dopamine receptor, ... | | Authors: | Yan, W, Shao, Z. | | Deposit date: | 2020-07-20 | | Release date: | 2021-03-03 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Ligand recognition and allosteric regulation of DRD1-Gs signaling complexes.
Cell, 184, 2021
|
|
7CKX
 
 | | Cryo-EM structure of A77636 bound dopamine receptor DRD1-Gs signaling complex | | Descriptor: | (1R,3S)-3-(1-adamantyl)-1-(aminomethyl)-3,4-dihydro-1H-isochromene-5,6-diol, CHOLESTEROL, D(1A) dopamine receptor, ... | | Authors: | Yan, W, Shao, Z. | | Deposit date: | 2020-07-20 | | Release date: | 2021-03-03 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | Ligand recognition and allosteric regulation of DRD1-Gs signaling complexes.
Cell, 184, 2021
|
|
