1EZF
 
 | | CRYSTAL STRUCTURE OF HUMAN SQUALENE SYNTHASE | | Descriptor: | FARNESYL-DIPHOSPHATE FARNESYLTRANSFERASE, N-{2-[TRANS-7-CHLORO-1-(2,2-DIMETHYL-PROPYL) -5-NAPHTHALEN-1-YL-2-OXO-1,2,3,5-TETRAHYDRO-BENZO[E] [1,4]OXAZEPIN-3-YL]-ACETYL}-ASPARTIC ACID | | Authors: | Pandit, J, Danley, D.E, Schulte, G.K, Mazzalupo, S.M, Pauly, T.A, Hayward, C.M, Hamanaka, E.S, Thompson, J.F, Harwood, H.J. | | Deposit date: | 2000-05-10 | | Release date: | 2000-10-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of human squalene synthase. A key enzyme in cholesterol biosynthesis.
J.Biol.Chem., 275, 2000
|
|
3ITM
 
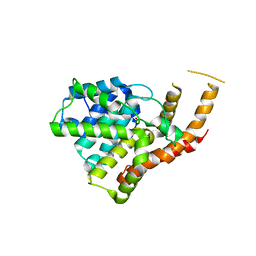 | | Catalytic domain of hPDE2A | | Descriptor: | ZINC ION, cGMP-dependent 3',5'-cyclic phosphodiesterase | | Authors: | Pandit, J. | | Deposit date: | 2009-08-28 | | Release date: | 2009-10-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Mechanism for the allosteric regulation of phosphodiesterase 2A deduced from the X-ray structure of a near full-length construct.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3IBJ
 
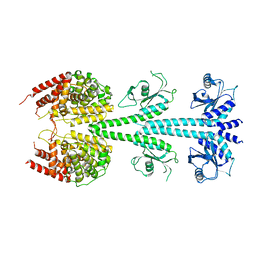 | | X-ray structure of PDE2A | | Descriptor: | MAGNESIUM ION, ZINC ION, cGMP-dependent 3',5'-cyclic phosphodiesterase | | Authors: | Pandit, J. | | Deposit date: | 2009-07-16 | | Release date: | 2009-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | Mechanism for the allosteric regulation of phosphodiesterase 2A deduced from the X-ray structure of a near full-length construct.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3ITU
 
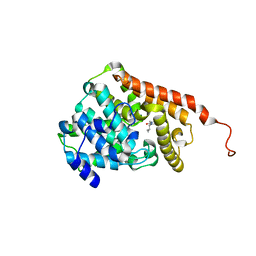 | | hPDE2A catalytic domain complexed with IBMX | | Descriptor: | 3-ISOBUTYL-1-METHYLXANTHINE, MAGNESIUM ION, ZINC ION, ... | | Authors: | Pandit, J. | | Deposit date: | 2009-08-28 | | Release date: | 2009-10-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Mechanism for the allosteric regulation of phosphodiesterase 2A deduced from the X-ray structure of a near full-length construct.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2OVY
 
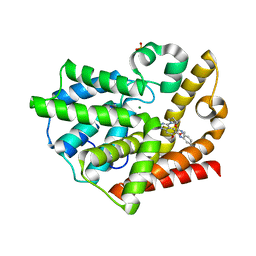 | | Crystal structure of the catalytic domain of rat phosphodiesterase 10A | | Descriptor: | 6,7-DIMETHOXY-4-[(3R)-3-(QUINOXALIN-2-YLOXY)PYRROLIDIN-1-YL]QUINAZOLINE, MAGNESIUM ION, Phosphodiesterase-10A, ... | | Authors: | Pandit, J, Marr, E.S. | | Deposit date: | 2007-02-15 | | Release date: | 2007-03-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of a series of 6,7-dimethoxy-4-pyrrolidylquinazoline PDE10A inhibitors
J.Med.Chem., 50, 2007
|
|
4JIB
 
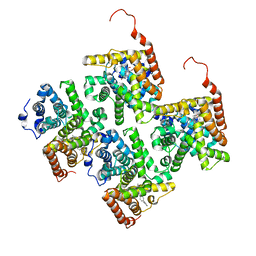 | | Crystal structure of of PDE2-inhibitor complex | | Descriptor: | 1-(2-hydroxyethyl)-3-(2-methylbutan-2-yl)-5-[4-(2-methyl-1H-imidazol-1-yl)phenyl]-6,7-dihydropyrazolo[4,3-e][1,4]diazepin-8(1H)-one, MAGNESIUM ION, ZINC ION, ... | | Authors: | Pandit, J. | | Deposit date: | 2013-03-05 | | Release date: | 2013-05-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Discovery of potent, selective, bioavailable phosphodiesterase 2 (PDE2) inhibitors active in an osteoarthritis pain model, Part I: Transformation of selective pyrazolodiazepinone phosphodiesterase 4 (PDE4) inhibitors into selective PDE2 inhibitors.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4LZ5
 
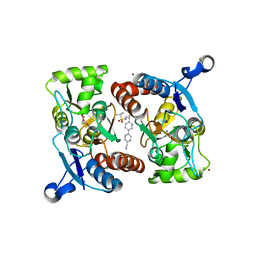 | |
4LZ8
 
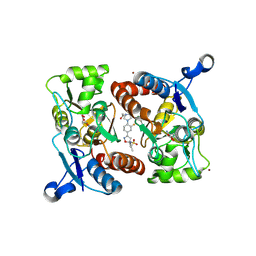 | |
4LZ7
 
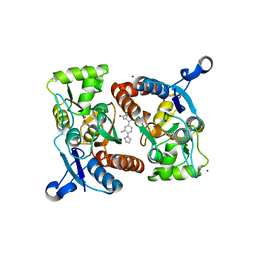 | |
5KRE
 
 | | Covalent inhibitor of LYPLAL1 | | Descriptor: | (2~{R})-2-phenylpiperidine-1-carbaldehyde, Lysophospholipase-like protein 1, NITRATE ION | | Authors: | Pandit, J. | | Deposit date: | 2016-07-07 | | Release date: | 2016-07-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of a Selective Covalent Inhibitor of Lysophospholipase-like 1 (LYPLAL1) as a Tool to Evaluate the Role of this Serine Hydrolase in Metabolism.
Acs Chem.Biol., 11, 2016
|
|
5EWJ
 
 | |
5EWM
 
 | |
5EWL
 
 | |
3CQU
 
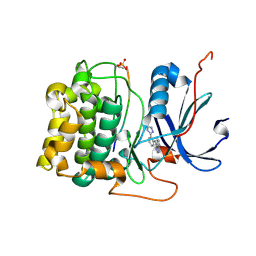 | | Crystal Structure of Akt-1 complexed with substrate peptide and inhibitor | | Descriptor: | Glycogen synthase kinase-3 beta, N-[2-(5-methyl-4H-1,2,4-triazol-3-yl)phenyl]-7H-pyrrolo[2,3-d]pyrimidin-4-amine, RAC-alpha serine/threonine-protein kinase | | Authors: | Pandit, J. | | Deposit date: | 2008-04-03 | | Release date: | 2008-05-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Synthesis and structure based optimization of novel Akt inhibitors
Bioorg.Med.Chem.Lett., 18, 2008
|
|
3CQW
 
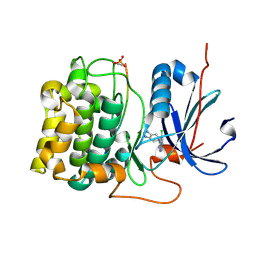 | | Crystal Structure of Akt-1 complexed with substrate peptide and inhibitor | | Descriptor: | 5-(5-chloro-7H-pyrrolo[2,3-d]pyrimidin-4-yl)-4,5,6,7-tetrahydro-1H-imidazo[4,5-c]pyridine, Glycogen synthase kinase-3 beta, MANGANESE (II) ION, ... | | Authors: | Pandit, J. | | Deposit date: | 2008-04-03 | | Release date: | 2008-05-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Synthesis and structure based optimization of novel Akt inhibitors
Bioorg.Med.Chem.Lett., 18, 2008
|
|
1HMC
 
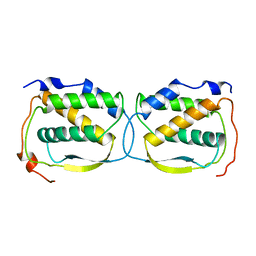 | | THREE-DIMENSIONAL STRUCTURE OF DIMERIC HUMAN RECOMBINANT MACROPHAGE COLONY STIMULATING FACTOR | | Descriptor: | HUMAN MACROPHAGE COLONY STIMULATING FACTOR | | Authors: | Bohm, A, Pandit, J, Jancarik, J, Halenbeck, R, Koths, K, Kim, S.-H. | | Deposit date: | 1993-12-22 | | Release date: | 1994-04-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Three-dimensional structure of dimeric human recombinant macrophage colony-stimulating factor.
Science, 258, 1992
|
|
4X48
 
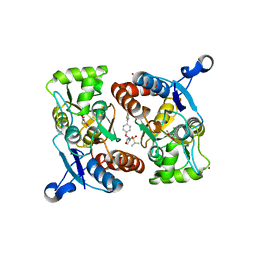 | | Crystal structure of GluR2 ligand-binding core | | Descriptor: | GLUTAMIC ACID, Glutamate receptor 2, N-{(3S,4S)-4-[4-(5-cyanothiophen-2-yl)phenoxy]tetrahydrofuran-3-yl}propane-2-sulfonamide, ... | | Authors: | Pandit, J. | | Deposit date: | 2014-12-02 | | Release date: | 2015-05-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | The Discovery and Characterization of the alpha-Amino-3-hydroxy-5-methyl-4-isoxazolepropionic Acid (AMPA) Receptor Potentiator N-{(3S,4S)-4-[4-(5-Cyano-2-thienyl)phenoxy]tetrahydrofuran-3-yl}propane-2-sulfonamide (PF-04958242).
J.Med.Chem., 58, 2015
|
|
6AX1
 
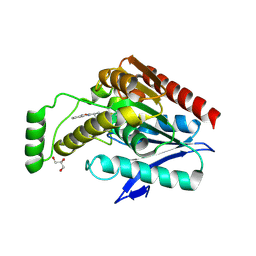 | | Structure of human monoacylglycerol lipase bound to a covalent inhibitor | | Descriptor: | 1,1,1,3,3,3-hexafluoropropan-2-yl 3-(3-phenyl-1,2,4-oxadiazol-5-yl)azetidine-1-carboxylate, GLYCEROL, Monoglyceride lipase | | Authors: | Pandit, J. | | Deposit date: | 2017-09-06 | | Release date: | 2017-12-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Azetidine and Piperidine Carbamates as Efficient, Covalent Inhibitors of Monoacylglycerol Lipase.
J. Med. Chem., 60, 2017
|
|
3MVH
 
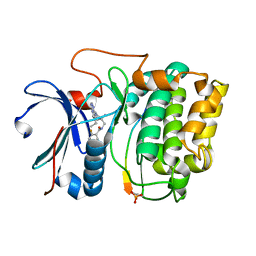 | | Crystal structure of Akt-1-inhibitor complexes | | Descriptor: | GSK3-beta peptide, MANGANESE (II) ION, N-{[(3S)-3-amino-1-(5-ethyl-7H-pyrrolo[2,3-d]pyrimidin-4-yl)pyrrolidin-3-yl]methyl}-2,4-difluorobenzamide, ... | | Authors: | Pandit, J. | | Deposit date: | 2010-05-04 | | Release date: | 2010-06-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Design of selective, ATP-competitive inhibitors of Akt.
J.Med.Chem., 53, 2010
|
|
3MV5
 
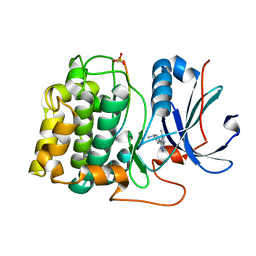 | | Crystal structure of Akt-1-inhibitor complexes | | Descriptor: | (3R)-1-(5-methyl-7H-pyrrolo[2,3-d]pyrimidin-4-yl)pyrrolidin-3-amine, GSK3-beta peptide, MANGANESE (II) ION, ... | | Authors: | Pandit, J. | | Deposit date: | 2010-05-03 | | Release date: | 2010-06-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Design of selective, ATP-competitive inhibitors of Akt.
J.Med.Chem., 53, 2010
|
|
3MVJ
 
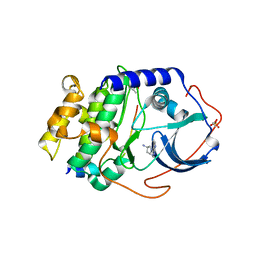 | | Human cyclic AMP-dependent protein kinase PKA inhibitor complex | | Descriptor: | (3R)-1-(5-methyl-7H-pyrrolo[2,3-d]pyrimidin-4-yl)pyrrolidin-3-amine, cAMP-dependent protein kinase catalytic subunit alpha, cAMP-dependent protein kinase inhibitor alpha | | Authors: | Pandit, J, Vajdos, F. | | Deposit date: | 2010-05-04 | | Release date: | 2010-06-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Design of selective, ATP-competitive inhibitors of Akt.
J.Med.Chem., 53, 2010
|
|
6BQ0
 
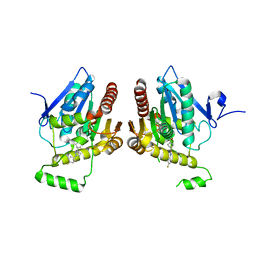 | | Structure of human monoacylglycerol lipase bound to a covalent inhibitor | | Descriptor: | 1-({(1R,5S,6r)-6-[1-(4-fluorophenyl)-1H-pyrazol-3-yl]-3-azabicyclo[3.1.0]hexane-3-carbonyl}oxy)pyrrolidine-2,5-dione, Monoglyceride lipase | | Authors: | Pandit, J. | | Deposit date: | 2017-11-27 | | Release date: | 2018-03-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of Trifluoromethyl Glycol Carbamates as Potent and Selective Covalent Monoacylglycerol Lipase (MAGL) Inhibitors for Treatment of Neuroinflammation.
J. Med. Chem., 61, 2018
|
|
4NPW
 
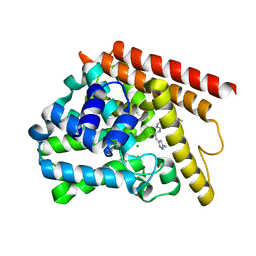 | | Crystal structure of human PDE1B bound to inhibitor 19A (7,8-dimethoxy-N-[(2S)-1-(3-methyl-1H-pyrazol-5-yl)propan-2-yl]quinazolin-4-amine) | | Descriptor: | 7,8-dimethoxy-N-[(2S)-1-(3-methyl-1H-pyrazol-5-yl)propan-2-yl]quinazolin-4-amine, Calcium/calmodulin-dependent 3',5'-cyclic nucleotide phosphodiesterase 1B, MAGNESIUM ION, ... | | Authors: | Pandit, J, Evdomikov, A, Mansour, M, Simons, S. | | Deposit date: | 2013-11-22 | | Release date: | 2014-07-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Small-molecule phosphodiesterase probes: discovery of potent and selective CNS-penetrable quinazoline inhibitors of PDE1
MEDCHEMCOMM, 2014
|
|
4NPV
 
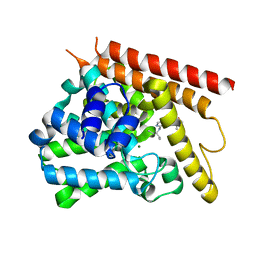 | | Crystal structure of human PDE1B bound to inhibitor 7A (6,7,8-trimethoxy-N-(pentan-3-yl)quinazolin-4-amine) | | Descriptor: | 6,7,8-trimethoxy-N-(pentan-3-yl)quinazolin-4-amine, Calcium/calmodulin-dependent 3',5'-cyclic nucleotide phosphodiesterase 1B, MAGNESIUM ION, ... | | Authors: | Pandit, J, Evdomikov, A, Mansour, M, Simons, S. | | Deposit date: | 2013-11-22 | | Release date: | 2014-07-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Small-molecule phosphodiesterase probes: discovery of potent and selective CNS-penetrable quinazoline inhibitors of PDE1
MEDCHEMCOMM, 2014
|
|
3QPP
 
 | | Structure of PDE10-inhibitor complex | | Descriptor: | 7-methoxy-4-[(3S)-3-phenylpiperidin-1-yl]-6-[2-(quinolin-2-yl)ethoxy]quinazoline, DIMETHYL SULFOXIDE, MAGNESIUM ION, ... | | Authors: | Pandit, J, Marr, E.S. | | Deposit date: | 2011-02-14 | | Release date: | 2011-06-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Use of Structure-Based Design to Discover a Potent, Selective, In Vivo Active Phosphodiesterase 10A Inhibitor Lead Series for the Treatment of Schizophrenia.
J.Med.Chem., 54, 2011
|
|
