2CQU
 
 | | Solution Structure of RSGI RUH-045, a Human Acyl-CoA Binding Protein | | Descriptor: | peroxisomal D3,D2-enoyl-CoA isomerase | | Authors: | Tsubota, Y, Ruhul Momen, A.Z.M, Onuki, H, Hirota, H, Saito, K, Koshiba, S, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-20 | | Release date: | 2005-11-20 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of RSGI RUH-045, a Human Acyl-CoA Binding Protein
To be Published
|
|
1PGR
 
 | | 2:2 COMPLEX OF G-CSF WITH ITS RECEPTOR | | Descriptor: | PROTEIN (G-CSF RECEPTOR), PROTEIN (GRANULOCYTE COLONY-STIMULATING FACTOR) | | Authors: | Aritomi, M, Kunishima, N, Okamoto, T, Kuroki, R, Ota, Y, Morikawa, K. | | Deposit date: | 1999-03-08 | | Release date: | 2000-03-08 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Atomic structure of the GCSF-receptor complex showing a new cytokine-receptor recognition scheme.
Nature, 401, 1999
|
|
1CD9
 
 | | 2:2 COMPLEX OF G-CSF WITH ITS RECEPTOR | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, PROTEIN (G-CSF RECEPTOR), PROTEIN (GRANULOCYTE COLONY-STIMULATING FACTOR) | | Authors: | Aritomi, M, Kunishima, N, Okamoto, T, Kuroki, R, Ota, Y, Morikawa, K. | | Deposit date: | 1999-03-08 | | Release date: | 2000-03-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Atomic structure of the GCSF-receptor complex showing a new cytokine-receptor recognition scheme.
Nature, 401, 1999
|
|
1GCF
 
 | | NMR STRUCTURE OF THE C-TERMINAL DOMAIN OF THE LIGAND-BINDING REGION OF MURINE GRANULOCYTE COLONY-STIMULATING FACTOR RECEPTOR, 12 STRUCTURES | | Descriptor: | GRANULOCYTE COLONY-STIMULATING FACTOR RECEPTOR | | Authors: | Yamasaki, K, Naito, S, Anaguchi, H, Ohkubo, T, Ota, Y. | | Deposit date: | 1997-04-10 | | Release date: | 1997-10-22 | | Last modified: | 2018-03-14 | | Method: | SOLUTION NMR | | Cite: | Solution structure of an extracellular domain containing the WSxWS motif of the granulocyte colony-stimulating factor receptor and its interaction with ligand.
Nat.Struct.Biol., 4, 1997
|
|
1CTO
 
 | | NMR STRUCTURE OF THE C-TERMINAL DOMAIN OF THE LIGAND-BINDING REGION OF MURINE GRANULOCYTE COLONY-STIMULATING FACTOR RECEPTOR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | GRANULOCYTE COLONY-STIMULATING FACTOR RECEPTOR | | Authors: | Yamasaki, K, Naito, S, Anaguchi, H, Ohkubo, T, Ota, Y. | | Deposit date: | 1996-09-25 | | Release date: | 1997-10-22 | | Last modified: | 2018-03-14 | | Method: | SOLUTION NMR | | Cite: | Solution structure of an extracellular domain containing the WSxWS motif of the granulocyte colony-stimulating factor receptor and its interaction with ligand.
Nat.Struct.Biol., 4, 1997
|
|
6L26
 
 | | Neutron crystal structure of the mutant green fluorescent protein (EGFP) | | Descriptor: | Green fluorescent protein, trideuteriooxidanium | | Authors: | Adachi, M, Shimizu, R, Shibazaki, C, Kagotani, Y, Ostermann, A, Schrader, T.E. | | Deposit date: | 2019-10-02 | | Release date: | 2020-04-08 | | Last modified: | 2023-11-22 | | Method: | NEUTRON DIFFRACTION (1.444 Å) | | Cite: | Direct Observation of the Protonation States in the Mutant Green Fluorescent Protein.
J Phys Chem Lett, 11, 2020
|
|
6L27
 
 | | X-ray crystal structure of the mutant green fluorescent protein | | Descriptor: | Green fluorescent protein | | Authors: | Adachi, M, Shimizu, R, Shibazaki, C, Kagotani, Y, Ostermann, A, Schrader, T.E. | | Deposit date: | 2019-10-02 | | Release date: | 2020-04-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (0.77 Å) | | Cite: | Direct Observation of the Protonation States in the Mutant Green Fluorescent Protein.
J Phys Chem Lett, 11, 2020
|
|
8GOS
 
 | | Crystal structure of fluorescent protein RasM | | Descriptor: | RasM | | Authors: | Adachi, M, Kagotani, Y, Shimizu, R. | | Deposit date: | 2022-08-25 | | Release date: | 2023-09-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.01 Å) | | Cite: | Beat-frequency-resolved two-dimensional electronic spectroscopy: disentangling vibrational coherences in artificial fluorescent proteins with sub-10-fs visible laser pulses.
Opt Express, 31, 2023
|
|
6A79
 
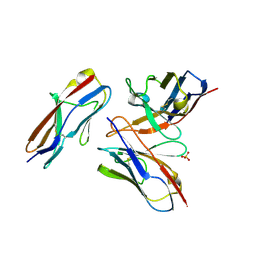 | | Crystal structure of the fifth immunoglobulin domain (Ig5) of human Robo1 in complex with the mutant scFv fragment (P103A) of murine monoclonal antibody B5209B | | Descriptor: | Heavy chain of the anti-human Robo1 antibody B5209B scFv, Light chain region of the anti-human Robo1 antibody B5209B scFv, Roundabout homolog 1, ... | | Authors: | Mizohata, E, Nakayama, T, Kado, Y, Yokota, Y, Inoue, T. | | Deposit date: | 2018-07-02 | | Release date: | 2019-01-30 | | Last modified: | 2019-03-20 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Affinity Improvement of a Cancer-Targeted Antibody through Alanine-Induced Adjustment of Antigen-Antibody Interface.
Structure, 27, 2019
|
|
3WII
 
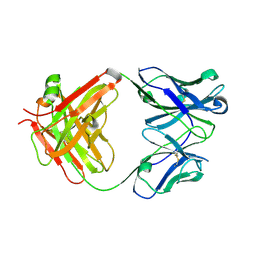 | | Crystal structure of the Fab fragment of B2212A, a murine monoclonal antibody specific for the third fibronectin domain (Fn3) of human ROBO1. | | Descriptor: | anti-human ROBO1 antibody B2212A Fab heavy chain, anti-human ROBO1 antibody B2212A Fab light chain | | Authors: | Nakayama, T, Mizohata, E, Yamashita, T, Nagatoishi, S, Nakakido, M, Iwanari, H, Mochizuki, Y, Kado, Y, Yokota, Y, Sato, R, Tsumoto, K, Fujitani, H, Kodama, T, Hamakubo, T, Inoue, T. | | Deposit date: | 2013-09-12 | | Release date: | 2015-01-21 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural features of interfacial tyrosine residue in ROBO1 fibronectin domain-antibody complex: Crystallographic, thermodynamic, and molecular dynamic analyses
Protein Sci., 24, 2015
|
|
3WIH
 
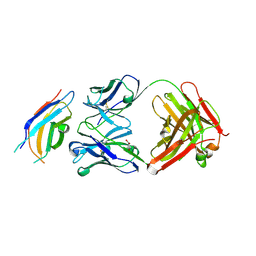 | | Crystal structure of the third fibronectin domain (Fn3) of human ROBO1 in complex with the Fab fragment of murine monoclonal antibody B2212A. | | Descriptor: | GLYCEROL, Roundabout homolog 1, anti-human ROBO1 antibody B2212A Fab heavy chain, ... | | Authors: | Nakayama, T, Mizohata, E, Yamashita, T, Nagatoishi, M, Iwanari, H, Mochizuki, Y, Kado, Y, Yokota, Y, Sato, R, Tsumoto, K, Fujitani, H, Kodama, T, Hamakubo, T, Inoue, T. | | Deposit date: | 2013-09-12 | | Release date: | 2015-01-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | Structural features of interfacial tyrosine residue in ROBO1 fibronectin domain-antibody complex: Crystallographic, thermodynamic, and molecular dynamic analyses
Protein Sci., 24, 2015
|
|
7C7D
 
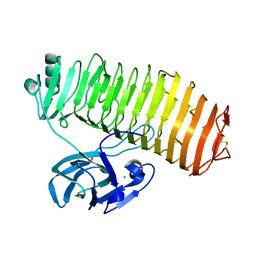 | | Crystal structure of the catalytic unit of thermostable GH87 alpha-1,3-glucanase from Streptomyces thermodiastaticus strain HF3-3 | | Descriptor: | CALCIUM ION, PENTAETHYLENE GLYCOL, alpha-1,3-glucanase | | Authors: | Itoh, T, Panti, N, Toyotake, Y, Hayashi, J, Suyotha, W, Yano, S, Wakayama, M, Hibi, T. | | Deposit date: | 2020-05-25 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Crystal structure of the catalytic unit of thermostable GH87 alpha-1,3-glucanase from Streptomyces thermodiastaticus strain HF3-3.
Biochem.Biophys.Res.Commun., 533, 2020
|
|
7F2X
 
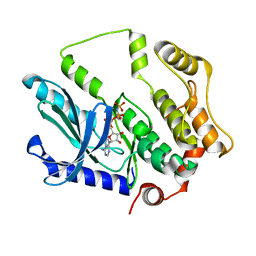 | | Crystal structure of MEK1 C121S mutant | | Descriptor: | MEK1 F11, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Fujioka, Y, Noda, N.N. | | Deposit date: | 2021-06-15 | | Release date: | 2022-06-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.007 Å) | | Cite: | Qualitative differences in disease-associated MEK mutants reveal molecular signatures and aberrant signaling-crosstalk in cancer.
Nat Commun, 13, 2022
|
|
6KG2
 
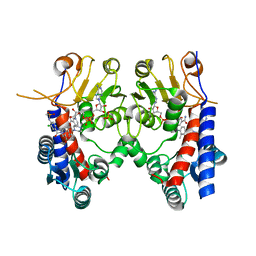 | | Human MTHFD2 in complex with Compound 18 | | Descriptor: | Bifunctional methylenetetrahydrofolate dehydrogenase/cyclohydrolase, mitochondrial, N-[2-chloranyl-4-[[7-methyl-8-(4-methylpiperazin-1-yl)-5-oxidanylidene-2,4-dihydro-1H-chromeno[3,4-c]pyridin-3-yl]carbonyl]phenyl]methanesulfonamide, ... | | Authors: | Suzuki, M, Matsui, Y, Ota, M, Kawai, J. | | Deposit date: | 2019-07-10 | | Release date: | 2019-11-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Discovery of a Potent, Selective, and Orally Available MTHFD2 Inhibitor (DS18561882) with in Vivo Antitumor Activity.
J.Med.Chem., 62, 2019
|
|
7BWW
 
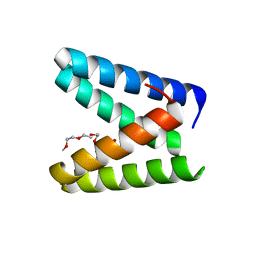 | | Structure of the engineered metallo-Diels-Alderase DA7 W16S | | Descriptor: | ACETIC ACID, BENZOIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Basler, S, Mori, T, Hilvert, D. | | Deposit date: | 2020-04-16 | | Release date: | 2021-04-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Efficient Lewis acid catalysis of an abiological reaction in a de novo protein scaffold.
Nat.Chem., 13, 2021
|
|
7Y8P
 
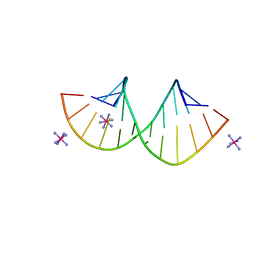 | | Crystal structure of 4'-selenoRNA duplex | | Descriptor: | COBALT HEXAMMINE(III), RNA (5'-R(*GP*GP*AP*(IKS)P*(ILK)P*(IKS)P*GP*AP*GP*UP*CP*C)-3') | | Authors: | Kondo, J, Minakawa, N, Ohta, M, Takahashi, H, Tarashima, N. | | Deposit date: | 2022-06-24 | | Release date: | 2023-05-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Synthesis and properties of fully-modified 4'-selenoRNA, an endonuclease-resistant RNA analog.
Bioorg.Med.Chem., 76, 2022
|
|
6YPI
 
 | | Structure of the engineered metallo-Diels-Alderase DA7 W16G,K58Q,L77R,T78R | | Descriptor: | 3-[BENZYL(DIMETHYL)AMMONIO]PROPANE-1-SULFONATE, BENZOIC ACID, DA7 W16G,K58Q,L77R,T78R, ... | | Authors: | Basler, S, Mori, T, Hilvert, D. | | Deposit date: | 2020-04-16 | | Release date: | 2021-04-28 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.479 Å) | | Cite: | Efficient Lewis acid catalysis of an abiological reaction in a de novo protein scaffold.
Nat.Chem., 13, 2021
|
|
6Z4T
 
 | |
6Z4R
 
 | |
4PK6
 
 | | Crystal structure of the indoleamine 2,3-dioxygenagse 1 (IDO1) complexed with imidazothiazole derivative | | Descriptor: | Indoleamine 2,3-dioxygenase 1, N-[2-(3-chlorophenyl)ethyl]-3-(4-methylphenyl)imidazo[2,1-b][1,3]thiazole-5-carboxamide, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Kohno, T, Tojo, S, Ishii, T, Kamioka, S. | | Deposit date: | 2014-05-13 | | Release date: | 2014-09-03 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Crystal Structures and Structure-Activity Relationships of Imidazothiazole Derivatives as IDO1 Inhibitors.
Acs Med.Chem.Lett., 5, 2014
|
|
4PK5
 
 | | Crystal structure of the indoleamine 2,3-dioxygenagse 1 (IDO1) complexed with Amg-1 | | Descriptor: | Indoleamine 2,3-dioxygenase 1, N-(1,3-benzodioxol-5-yl)-2-{[5-(4-methylphenyl)[1,3]thiazolo[2,3-c][1,2,4]triazol-3-yl]sulfanyl}acetamide, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Kohno, T, Tojo, S, Ishii, T. | | Deposit date: | 2014-05-13 | | Release date: | 2014-09-03 | | Last modified: | 2020-01-29 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Crystal Structures and Structure-Activity Relationships of Imidazothiazole Derivatives as IDO1 Inhibitors.
Acs Med.Chem.Lett., 5, 2014
|
|
6LCS
 
 | | Crystal structure of 73MuL9 Fv-clasp fragment in complex with GA-pyridine analogue | | Descriptor: | (2~{S})-6-[4-(hydroxymethyl)-3-oxidanyl-pyridin-1-ium-1-yl]-2-(phenylmethoxycarbonylamino)hexanoic acid, PHOSPHATE ION, VH-SARAH, ... | | Authors: | Nakamura, T, Takagi, J, Yamagata, Y, Morioka, H. | | Deposit date: | 2019-11-19 | | Release date: | 2020-11-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Molecular recognition of a single-chain Fv antibody specific for GA-pyridine, an advanced glycation end-product (AGE), elucidated using biophysical techniques and synthetic antigen analogues.
J.Biochem., 170, 2021
|
|
8I2Q
 
 | | Beijerinckia indica beta-fructosyltransferase variant H395R/F473Y | | Descriptor: | Beta-fructosyltransferase, GLYCEROL | | Authors: | Tonozuka, T. | | Deposit date: | 2023-01-15 | | Release date: | 2023-06-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Characterization and alteration of product specificity of Beijerinckia indica subsp. indica beta-fructosyltransferase.
Biosci.Biotechnol.Biochem., 87, 2023
|
|
8I2R
 
 | |
1C8N
 
 | | TOBACCO NECROSIS VIRUS | | Descriptor: | CALCIUM ION, COAT PROTEIN | | Authors: | Oda, Y, Fukuyama, K. | | Deposit date: | 2000-05-20 | | Release date: | 2000-08-30 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of tobacco necrosis virus at 2.25 A resolution.
J.Mol.Biol., 300, 2000
|
|
