1Q1U
 
 | | Crystal structure of human FHF1b (FGF12b) | | Descriptor: | SULFATE ION, fibroblast growth factor homologous factor 1 | | Authors: | Olsen, S.K, Garbi, M, Zampieri, N, Eliseenkova, A.V, Ornitz, D.M, Goldfarb, M, Mohammadi, M. | | Deposit date: | 2003-07-22 | | Release date: | 2003-08-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Fibroblast growth factor (FGF) homologous factors share structural but not functional homology with FGFs
J.Biol.Chem., 278, 2003
|
|
4II3
 
 | |
2PSM
 
 | | Crystal structure of Interleukin 15 in complex with Interleukin 15 receptor alpha | | Descriptor: | BENZAMIDINE, Interleukin-15, Interleukin-15 receptor alpha chain | | Authors: | Olsen, S.K, Murayama, K, Kishishita, S, Kukimoto-Niino, M, Terada, T, Shirouzu, M, Ota, N, Kanagawa, O, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-05-07 | | Release date: | 2007-11-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Crystal Structure of the Interleukin-15{middle dot}Interleukin-15 Receptor {alpha} Complex: INSIGHTS INTO TRANS AND CIS PRESENTATION
J.Biol.Chem., 282, 2007
|
|
1RY7
 
 | | Crystal Structure of the 3 Ig form of FGFR3c in complex with FGF1 | | Descriptor: | Fibroblast growth factor receptor 3, Heparin-binding growth factor 1 | | Authors: | Olsen, S.K, Ibrahimi, O.A, Raucci, A, Zhang, F, Eliseenkova, A.V, Yayon, A, Basilico, C, Linhardt, R.J, Schlessinger, J, Mohammadi, M. | | Deposit date: | 2003-12-19 | | Release date: | 2004-02-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Insights into the molecular basis for fibroblast growth factor receptor autoinhibition and ligand-binding promiscuity.
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
4II2
 
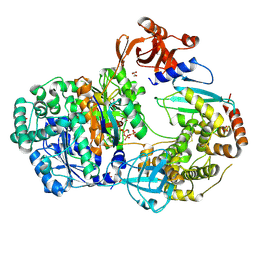 | | Crystal structure of Ubiquitin activating enzyme 1 (Uba1) in complex with the Ub E2 Ubc4, ubiquitin, and ATP/Mg | | Descriptor: | 1,2-ETHANEDIOL, 2-(2-METHOXYETHOXY)ETHANOL, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Olsen, S.K, Lima, C.D. | | Deposit date: | 2012-12-19 | | Release date: | 2013-02-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of a ubiquitin E1-E2 complex: insights to E1-E2 thioester transfer.
Mol.Cell, 49, 2013
|
|
8FY0
 
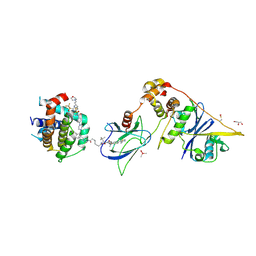 | | E3:PROTAC:target ternary complex structure (VCB/753b/BCL-xL) | | Descriptor: | Bcl-2-like protein 1, CACODYLIC ACID, Elongin-B, ... | | Authors: | Olsen, S.K, Nayak, D, Lv, D, Yuan, Y, Zhang, P, Hu, W, Lv, Z, Sung, P, Hromas, R, Zheng, G, Zhou, D. | | Deposit date: | 2023-01-25 | | Release date: | 2024-04-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Development and crystal structures of a potent second-generation dual degrader of BCL-2 and BCL-xL.
Nat Commun, 15, 2024
|
|
5KNL
 
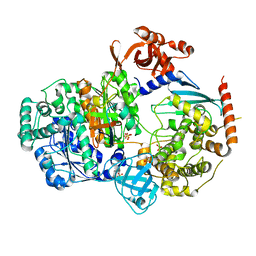 | | Crystal structure of S. pombe ubiquitin E1 (Uba1) in complex with Ubc15 and ubiquitin | | Descriptor: | SULFATE ION, Ubiquitin, Ubiquitin-activating enzyme E1 1, ... | | Authors: | Olsen, S.K, Lv, Z, Yuan, L, Williams, K. | | Deposit date: | 2016-06-28 | | Release date: | 2017-02-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | S. pombe Uba1-Ubc15 Structure Reveals a Novel Regulatory Mechanism of Ubiquitin E2 Activity.
Mol. Cell, 65, 2017
|
|
7LFV
 
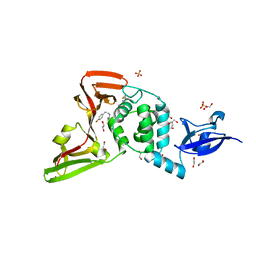 | |
7LFU
 
 | |
7SOL
 
 | | Crystal Structures of the bispecific ubiquitin/FAT10 activating enzyme, Uba6 | | Descriptor: | ADENOSINE MONOPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, Ubiquitin, ... | | Authors: | Olsen, S.K, Gao, F, Lv, Z. | | Deposit date: | 2021-10-31 | | Release date: | 2022-11-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.25000644 Å) | | Cite: | Crystal structures reveal catalytic and regulatory mechanisms of the dual-specificity ubiquitin/FAT10 E1 enzyme Uba6.
Nat Commun, 13, 2022
|
|
6NYA
 
 | | Crystal Structure of ubiquitin E1 (Uba1) in complex with Ubc3 (Cdc34) and ubiquitin | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Olsen, S.K, Williams, K.M, Atkison, J.H. | | Deposit date: | 2019-02-11 | | Release date: | 2019-08-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.065 Å) | | Cite: | Structural insights into E1 recognition and the ubiquitin-conjugating activity of the E2 enzyme Cdc34.
Nat Commun, 10, 2019
|
|
6NYO
 
 | | Crystal structure of a human Cdc34-ubiquitin thioester mimetic | | Descriptor: | 1,2-ETHANEDIOL, 4,5-dideoxy-5-(3',5'-dichlorobiphenyl-4-yl)-4-[(methoxyacetyl)amino]-L-arabinonic acid, PHOSPHATE ION, ... | | Authors: | Olsen, S.K, Williams, K.M, Atkison, J.H. | | Deposit date: | 2019-02-11 | | Release date: | 2019-08-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.502 Å) | | Cite: | Structural insights into E1 recognition and the ubiquitin-conjugating activity of the E2 enzyme Cdc34.
Nat Commun, 10, 2019
|
|
6O82
 
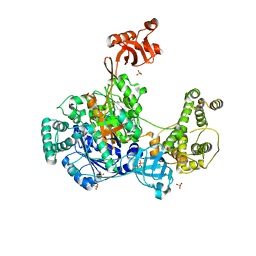 | | S. pombe ubiquitin E1 complex with a ubiquitin-AMP mimic | | Descriptor: | 1,2-ETHANEDIOL, 5'-deoxy-5'-(sulfamoylamino)adenosine, MAGNESIUM ION, ... | | Authors: | Olsen, S.K, Lima, C.D. | | Deposit date: | 2019-03-08 | | Release date: | 2019-06-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.604 Å) | | Cite: | Structural basis for adenylation and thioester bond formation in the ubiquitin E1.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6NYD
 
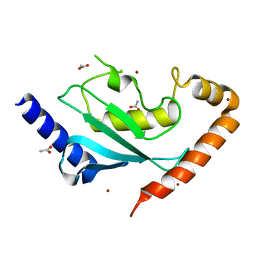 | |
2FDB
 
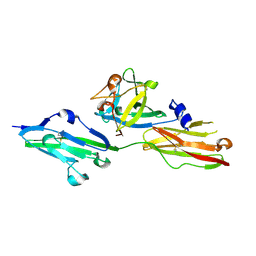 | |
8GBJ
 
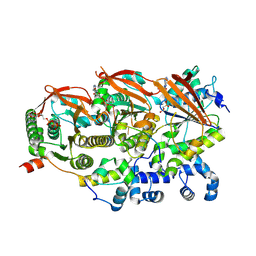 | | Cryo-EM structure of a human BCDX2/ssDNA complex | | Descriptor: | DNA (5'-D(P*CP*CP*CP*CP*CP*C)-3'), DNA repair protein RAD51 homolog 2, DNA repair protein RAD51 homolog 3, ... | | Authors: | Jia, L, Wasmuth, E.V, Ruben, E.A, Sung, P, Rawal, Y, Greene, E.C, Meir, A, Olsen, S.K. | | Deposit date: | 2023-02-26 | | Release date: | 2023-06-21 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.11 Å) | | Cite: | Structural insights into BCDX2 complex function in homologous recombination.
Nature, 619, 2023
|
|
6WUU
 
 | |
6WX4
 
 | |
8FY2
 
 | | E3:PROTAC:target ternary complex structure (VCB/WH244/BCL-2) | | Descriptor: | Apoptosis regulator Bcl-2, Elongin-B, Elongin-C, ... | | Authors: | Nayak, D, Lv, D, Yuan, Y, Zhang, P, Hu, W, Ruben, E, Lv, Z, Sung, P, Hromas, R, Zheng, G, Zhou, D, Olsen, S.K. | | Deposit date: | 2023-01-25 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Development and crystal structures of a potent second-generation dual degrader of BCL-2 and BCL-xL.
Nat Commun, 15, 2024
|
|
8FY1
 
 | | E3:PROTAC:target ternary complex structure (VCB/753b/BCL-2) | | Descriptor: | Apoptosis regulator Bcl-2, Elongin-B, Elongin-C, ... | | Authors: | Nayak, D, Lv, D, Yuan, Y, Zhang, P, Hu, W, Lv, Z, Sung, P, Hromas, R, Zheng, G, Zhou, D, Olsen, S.K. | | Deposit date: | 2023-01-25 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Development and crystal structures of a potent second-generation dual degrader of BCL-2 and BCL-xL.
Nat Commun, 15, 2024
|
|
8FAZ
 
 | | Cryo-EM structure of the human BCDX2 complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA repair protein RAD51 homolog 2, DNA repair protein RAD51 homolog 3, ... | | Authors: | Jia, L, Wasmuth, E.V, Ruben, E.A, Sung, P, Rawal, Y, Greene, E.C, Meir, A, Olsen, S.K. | | Deposit date: | 2022-11-29 | | Release date: | 2023-06-21 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Structural insights into BCDX2 complex function in homologous recombination.
Nature, 619, 2023
|
|
7K5J
 
 | | Structure of an E1-E2-ubiquitin thioester mimetic | | Descriptor: | ADENOSINE MONOPHOSPHATE, Ubiquitin, Ubiquitin-activating enzyme E1 1, ... | | Authors: | Yuan, L, Lv, Z, Olsen, S.K. | | Deposit date: | 2020-09-16 | | Release date: | 2021-04-28 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (3.42 Å) | | Cite: | Crystal structures of an E1-E2-ubiquitin thioester mimetic reveal molecular mechanisms of transthioesterification.
Nat Commun, 12, 2021
|
|
5IZ2
 
 | | Crystal structure of the N. clavipes spidroin NTD at pH 6.5 | | Descriptor: | Major ampullate spidroin 1A, Major ampullate spidroin 1A (Partial C-terminus) | | Authors: | Atkison, J.H, Olsen, S.K. | | Deposit date: | 2016-03-24 | | Release date: | 2016-07-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Crystal Structure of the Nephila clavipes Major Ampullate Spidroin 1A N-terminal Domain Reveals Plasticity at the Dimer Interface.
J.Biol.Chem., 291, 2016
|
|
9BBU
 
 | | SARS-CoV-2 Mpro in complex with compound 6h inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 3C-like proteinase nsp5, N~2~-[4-(5-chloropyridin-3-yl)benzoyl]-N-{(1Z,2S)-1-imino-3-[(3R)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-leucinamide | | Authors: | Nayak, A, Afsar, M, Nayak, D, Olsen, S.K. | | Deposit date: | 2024-04-07 | | Release date: | 2025-04-16 | | Last modified: | 2025-05-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | SARS-CoV-2 Mpro in complex with compound 6h inhibitor
To Be Published
|
|
9BBR
 
 | | SARS-CoV-2 Mpro in complex with compound 6b inhibitor | | Descriptor: | 3C-like proteinase nsp5, 4'-fluoro-N-[(2S)-1-({(2R)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}amino)-4-methyl-1-oxopentan-2-yl][1,1'-biphenyl]-4-carboxamide | | Authors: | Afsar, M, Bury, P, Olsen, S.K. | | Deposit date: | 2024-04-07 | | Release date: | 2025-04-16 | | Last modified: | 2025-05-14 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | SARS-CoV-2 Mpro in complex with compound 6b inhibitor
To Be Published
|
|
