7M74
 
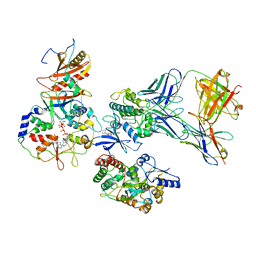 | | ATP-bound AMP-activated protein kinase | | Descriptor: | 5'-AMP-activated protein kinase catalytic subunit alpha-1, 5'-AMP-activated protein kinase subunit beta-2, 5'-AMP-activated protein kinase subunit gamma-1, ... | | Authors: | Yan, Y, Mukherjee, S, Harikumar, K.G, Strutzenberg, T, Zhou, X.E, Powell, S.K, Xu, T, Sheldon, R, Lamp, J, Brunzelle, J.S, Radziwon, K, Ellis, A, Novick, S.J, Vega, I.E, Jones, R, Miller, L.J, Xu, H.E, Griffin, P.R, Kossiakoff, A.A, Melcher, K. | | Deposit date: | 2021-03-26 | | Release date: | 2021-12-15 | | Last modified: | 2021-12-22 | | Method: | ELECTRON MICROSCOPY (3.93 Å) | | Cite: | Structure of an AMPK complex in an inactive, ATP-bound state.
Science, 373, 2021
|
|
8D1B
 
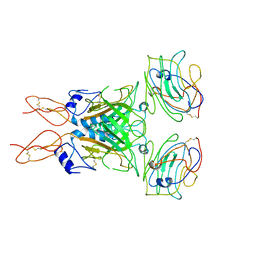 | |
6C9H
 
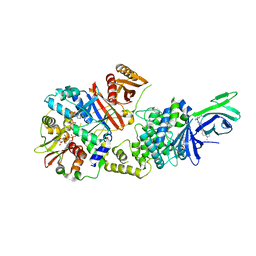 | | non-phosphorylated AMP-activated protein kinase bound to pharmacological activator R734 | | Descriptor: | 5'-AMP-activated protein kinase catalytic subunit alpha-1, 5'-AMP-activated protein kinase subunit beta-1, 5'-AMP-activated protein kinase subunit gamma-1, ... | | Authors: | Yan, Y, Zhou, X.E, Novick, S, Shaw, S.J, Li, Y, Hitoshi, Y, Brunzelle, J.S, Griffin, P.R, Xu, H.E, Melcher, K. | | Deposit date: | 2018-01-26 | | Release date: | 2018-11-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structures of AMP-activated protein kinase bound to novel pharmacological activators in phosphorylated, non-phosphorylated, and nucleotide-free states.
J. Biol. Chem., 294, 2019
|
|
6C9J
 
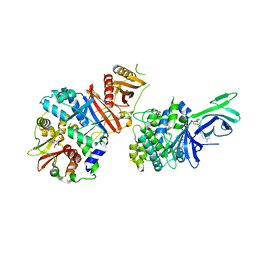 | | AMP-activated protein kinase bound to pharmacological activator R734 | | Descriptor: | 5'-AMP-activated protein kinase catalytic subunit alpha-1, 5'-AMP-activated protein kinase subunit beta-1, 5'-AMP-activated protein kinase subunit gamma-1, ... | | Authors: | Yan, Y, Zhou, X.E, Novick, S, Shaw, S.J, Li, Y, Brunzelle, J.S, Hitoshi, Y, Griffin, P.R, Xu, H.E, Melcher, K. | | Deposit date: | 2018-01-26 | | Release date: | 2018-11-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Structures of AMP-activated protein kinase bound to novel pharmacological activators in phosphorylated, non-phosphorylated, and nucleotide-free states.
J. Biol. Chem., 294, 2019
|
|
6C9F
 
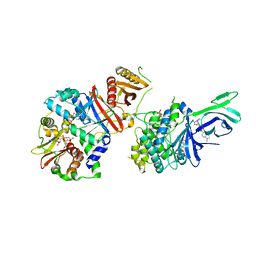 | | AMP-activated protein kinase bound to pharmacological activator R734 | | Descriptor: | 5'-AMP-activated protein kinase catalytic subunit alpha-1,5'-AMP-activated protein kinase catalytic subunit alpha-1, 5'-AMP-activated protein kinase subunit beta-1, 5'-AMP-activated protein kinase subunit gamma-1, ... | | Authors: | Yan, Y, Zhou, X.E, Novick, S, Shaw, S.J, Li, Y, Hitoshi, Y, Brunzelle, J.S, Griffin, P.R, Xu, H.E, Melcher, K. | | Deposit date: | 2018-01-26 | | Release date: | 2018-11-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.924 Å) | | Cite: | Structures of AMP-activated protein kinase bound to novel pharmacological activators in phosphorylated, non-phosphorylated, and nucleotide-free states.
J. Biol. Chem., 294, 2019
|
|
6C9G
 
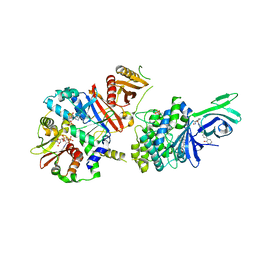 | | AMP-activated protein kinase bound to pharmacological activator R739 | | Descriptor: | 5'-AMP-activated protein kinase catalytic subunit alpha-1,5'-AMP-activated protein kinase catalytic subunit alpha-1, 5'-AMP-activated protein kinase subunit beta-1, 5'-AMP-activated protein kinase subunit gamma-1, ... | | Authors: | Yan, Y, Zhou, X.E, Novick, S, Shaw, S.J, Li, Y, Hitoshi, Y, Brunzelle, J.S, Griffin, P.R, Xu, H.E, Melcher, K. | | Deposit date: | 2018-01-26 | | Release date: | 2018-11-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structures of AMP-activated protein kinase bound to novel pharmacological activators in phosphorylated, non-phosphorylated, and nucleotide-free states.
J. Biol. Chem., 294, 2019
|
|
6N9G
 
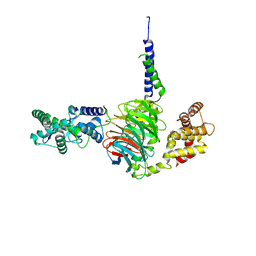 | | Crystal Structure of RGS7-Gbeta5 dimer | | Descriptor: | Guanine nucleotide-binding protein subunit beta-5, Regulator of G-protein signaling 7 | | Authors: | Patil, D.N, Rangarajan, E, Izard, T, Martemyanov, K.A. | | Deposit date: | 2018-12-03 | | Release date: | 2019-01-09 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.129 Å) | | Cite: | Structural organization of a major neuronal G protein regulator, the RGS7-G beta 5-R7BP complex.
Elife, 7, 2018
|
|
6NWS
 
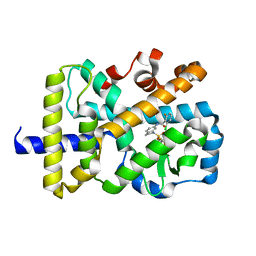 | | RORgamma Ligand Binding Domain | | Descriptor: | 2-chloro-6-fluoro-N-(1-{[3-(trifluoromethyl)phenyl]sulfonyl}-2,3-dihydro-1H-indol-6-yl)benzamide, Nuclear receptor ROR-gamma | | Authors: | Strutzenberg, T.S, Park, H.J, Griffin, P.R. | | Deposit date: | 2019-02-07 | | Release date: | 2019-07-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | HDX-MS reveals structural determinants for ROR gamma hyperactivation by synthetic agonists.
Elife, 8, 2019
|
|
6NWU
 
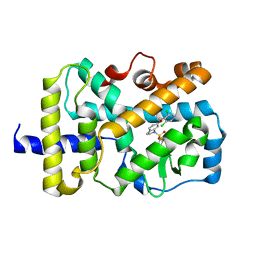 | | RORgamma Ligand Binding Domain | | Descriptor: | 6-[(3,5-dichloropyridin-4-yl)methoxy]-1-{[3-(trifluoromethyl)phenyl]sulfonyl}-2,3-dihydro-1H-indole, Nuclear receptor ROR-gamma | | Authors: | Strutzenberg, T.S, Park, H, Griffin, P.R. | | Deposit date: | 2019-02-07 | | Release date: | 2019-07-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | HDX-MS reveals structural determinants for ROR gamma hyperactivation by synthetic agonists.
Elife, 8, 2019
|
|
6NWT
 
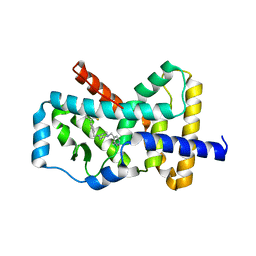 | | RORgamma Ligand Binding Domain | | Descriptor: | 1,1,1,3,3,3-hexafluoro-2-[2-fluoro-4'-({4-[(pyridin-4-yl)methyl]piperazin-1-yl}methyl)[1,1'-biphenyl]-4-yl]propan-2-ol, Nuclear receptor ROR-gamma | | Authors: | Strutzenberg, T.S, Park, H, Griffin, P.R. | | Deposit date: | 2019-02-07 | | Release date: | 2019-07-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | HDX-MS reveals structural determinants for ROR gamma hyperactivation by synthetic agonists.
Elife, 8, 2019
|
|
6E9F
 
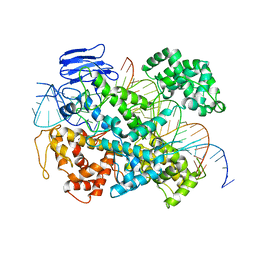 | | EsCas13d-crRNA-target RNA ternary complex | | Descriptor: | EsCas13d, MAGNESIUM ION, RNA (27-MER), ... | | Authors: | Zhang, C, Lyumkis, D. | | Deposit date: | 2018-08-01 | | Release date: | 2018-10-03 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural Basis for the RNA-Guided Ribonuclease Activity of CRISPR-Cas13d.
Cell, 175, 2018
|
|
6E9E
 
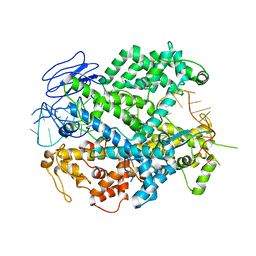 | | EsCas13d-crRNA binary complex | | Descriptor: | EsCas13d, MAGNESIUM ION, crRNA (52-MER) | | Authors: | Zhang, C, Lyumkis, D. | | Deposit date: | 2018-08-01 | | Release date: | 2018-10-03 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural Basis for the RNA-Guided Ribonuclease Activity of CRISPR-Cas13d.
Cell, 175, 2018
|
|
7ZC9
 
 | | Human Pikachurin/EGFLAM C-terminal Laminin-G domain (LG3) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Pikachurin, SULFATE ION | | Authors: | Pantalone, S, Forneris, F. | | Deposit date: | 2022-03-26 | | Release date: | 2023-07-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the photoreceptor synaptic assembly of the extracellular matrix protein pikachurin with the orphan receptor GPR179.
Sci.Signal., 16, 2023
|
|
7ZCB
 
 | | Human Pikachurin/EGFLAM N-terminal Fibronectin-III (1-2) domains | | Descriptor: | CHLORIDE ION, Pikachurin, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Pantalone, S, Savino, S, Viti, L.V, Forneris, F. | | Deposit date: | 2022-03-26 | | Release date: | 2023-07-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the photoreceptor synaptic assembly of the extracellular matrix protein pikachurin with the orphan receptor GPR179.
Sci.Signal., 16, 2023
|
|
7KSM
 
 | | Human mitochondrial LONP1 with endogenous substrate | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Lon protease homolog, ... | | Authors: | Shin, M, Watson, E.R, Song, A.S, Mindrebo, J.T, Novick, S.R, Griffin, P, Wiseman, R.L, Lander, G.C. | | Deposit date: | 2020-11-23 | | Release date: | 2020-12-02 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structures of the human LONP1 protease reveal regulatory steps involved in protease activation.
Nat Commun, 12, 2021
|
|
7KRZ
 
 | | Human mitochondrial LONP1 in complex with Bortezomib | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Endogenous co-purified substrate, ... | | Authors: | Shin, M, Watson, E.R, Song, A.S, Mindrebo, J.T, Novick, S.R, Griffin, P, Wiseman, R.L, Lander, G.C. | | Deposit date: | 2020-11-20 | | Release date: | 2021-02-24 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structures of the human LONP1 protease reveal regulatory steps involved in protease activation.
Nat Commun, 12, 2021
|
|
7KSL
 
 | | Substrate-free human mitochondrial LONP1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Lon protease homolog, mitochondrial | | Authors: | Shin, M, Watson, E.R, Song, A.S, Mindrebo, J.T, Novick, S.R, Griffin, P, Wiseman, R.L, Lander, G.C. | | Deposit date: | 2020-11-23 | | Release date: | 2020-12-09 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structures of the human LONP1 protease reveal regulatory steps involved in protease activation.
Nat Commun, 12, 2021
|
|
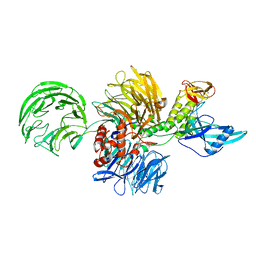
8D7Y
 
 | |
8D7X
 
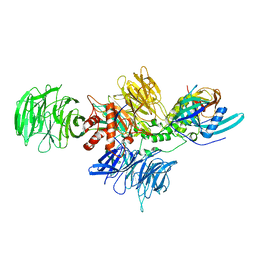 | |
4R6S
 
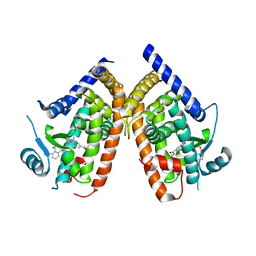 | | Crystal structure of PPARgammma in complex with SR1663 | | Descriptor: | 4'-[(2,3-dimethyl-5-{[(1R)-1-(4-nitrophenyl)ethyl]carbamoyl}-1H-indol-1-yl)methyl]biphenyl-2-carboxylic acid, Peroxisome proliferator-activated receptor gamma | | Authors: | Marciano, D.P, Griffin, P.R, Bruning, J.B. | | Deposit date: | 2014-08-26 | | Release date: | 2015-07-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Pharmacological repression of PPAR gamma promotes osteogenesis.
Nat Commun, 6, 2015
|
|
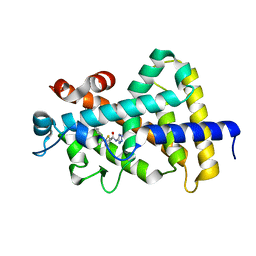
5V39
 
 | |
7JHG
 
 | | Cryo-EM structure of ATP-bound fully inactive AMPK in complex with Dorsomorphin (Compound C) and Fab-nanobody | | Descriptor: | 5'-AMP-activated protein kinase catalytic subunit alpha-1, 5'-AMP-activated protein kinase subunit beta-2, 5'-AMP-activated protein kinase subunit gamma-1, ... | | Authors: | Yan, Y, Murkherjee, S, Zhou, X.E, Xu, T.H, Xu, H.E, Kossiakoff, A.A, Melcher, K. | | Deposit date: | 2020-07-20 | | Release date: | 2021-07-21 | | Last modified: | 2021-12-15 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Structure of an AMPK complex in an inactive, ATP-bound state.
Science, 373, 2021
|
|
7JHH
 
 | | Cryo-EM structure of ATP-bound fully inactive AMPK in complex with Fab and nanobody | | Descriptor: | 5'-AMP-activated protein kinase catalytic subunit alpha-1, 5'-AMP-activated protein kinase subunit beta-2, 5'-AMP-activated protein kinase subunit gamma-1, ... | | Authors: | Yan, Y, Murkherjee, S, Zhou, X.E, Xu, T.H, Xu, H.E, Kossiakoff, A.A, Melcher, K. | | Deposit date: | 2020-07-20 | | Release date: | 2021-07-21 | | Last modified: | 2021-12-15 | | Method: | ELECTRON MICROSCOPY (3.92 Å) | | Cite: | Structure of an AMPK complex in an inactive, ATP-bound state.
Science, 373, 2021
|
|
7JIJ
 
 | | ATP-bound AMP-activated protein kinase | | Descriptor: | 5'-AMP-activated protein kinase catalytic subunit alpha-1, 5'-AMP-activated protein kinase subunit beta-2, 5'-AMP-activated protein kinase subunit gamma-1, ... | | Authors: | Yan, Y, Zhou, X.E, Powell, K, Xu, T, Brunzelle, J.S, Xu, H.X, Melcher, K. | | Deposit date: | 2020-07-23 | | Release date: | 2021-07-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (5.5 Å) | | Cite: | Structure of an AMPK complex in an inactive, ATP-bound state.
Science, 373, 2021
|
|
6D3E
 
 | | PPARg LBD in Complex with SR1988 | | Descriptor: | 1-[(2,4-difluorophenyl)methyl]-2,3-dimethyl-N-[(1R)-1-phenylpropyl]-1H-indole-5-carboxamide, Peroxisome proliferator-activated receptor gamma | | Authors: | Frkic, R.L, Bruning, J.B. | | Deposit date: | 2018-04-15 | | Release date: | 2019-02-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.395 Å) | | Cite: | Structural and Dynamic Elucidation of a Non-acid PPARgammaPartial Agonist: SR1988.
Nucl Receptor Res, 5, 2018
|
|
