4WUB
 
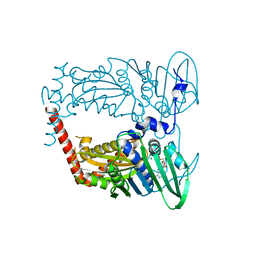 | | N-terminal 43 kDa fragment of the E. coli DNA gyrase B subunit grown from 100 mM KCl condition | | Descriptor: | CHLORIDE ION, DNA gyrase subunit B, MAGNESIUM ION, ... | | Authors: | Hearnshaw, S.J, Chung, T.T, Stevenson, C.E.M, Maxwell, A, Lawson, D.M. | | Deposit date: | 2014-10-31 | | Release date: | 2015-04-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The role of monovalent cations in the ATPase reaction of DNA gyrase
Acta Crystallogr.,Sect.D, 71, 2015
|
|
6FQV
 
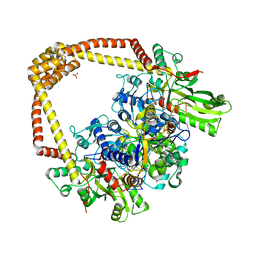 | | 2.60A BINARY COMPLEX OF S.AUREUS GYRASE with UNCLEAVED DNA | | Descriptor: | DNA (5'-D(*GP*AP*GP*CP*GP*TP*AP*CP*GP*GP*CP*CP*GP*TP*AP*CP*GP*CP*TP*T)-3'), DNA gyrase subunit A, DNA gyrase subunit B,DNA gyrase subunit B, ... | | Authors: | Bax, B.D, Germe, T, Basque, E, Maxwell, A. | | Deposit date: | 2018-02-14 | | Release date: | 2018-04-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A new class of antibacterials, the imidazopyrazinones, reveal structural transitions involved in DNA gyrase poisoning and mechanisms of resistance.
Nucleic Acids Res., 46, 2018
|
|
9FOY
 
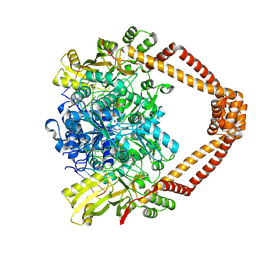 | | Ternary complex of a Mycobacterium tuberculosis DNA gyrase core fusion with DNA and the inhibitor AMK32b | | Descriptor: | (1~{R})-2-[4-[[4-bromanyl-3,5-bis(fluoranyl)phenyl]methylamino]cyclohexyl]-1-(6-methoxy-1,5-naphthyridin-4-yl)ethanol, (1~{S})-2-[4-[[4-bromanyl-3,5-bis(fluoranyl)phenyl]methylamino]cyclohexyl]-1-(6-methoxy-1,5-naphthyridin-4-yl)ethanol, DNA (5'-D(*AP*GP*CP*CP*GP*TP*AP*G)-3'), ... | | Authors: | Kokot, M, Hrast, M, Feng, L, Mitchenall, L.A, Lawson, D.M, Maxwell, A, Minovski, N, Anderluh, M. | | Deposit date: | 2024-06-12 | | Release date: | 2024-08-07 | | Last modified: | 2025-02-19 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Aspects of Mycobacterium tuberculosis DNA Gyrase Targeted by Novel Bacterial Topoisomerase Inhibitors.
Acs Med.Chem.Lett., 15, 2024
|
|
8BN6
 
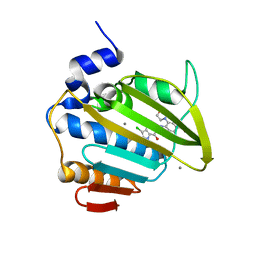 | | Pseudomonas aeruginosa DNA gyrase B 24kDa ATPase subdomain complexed with EBL3021 | | Descriptor: | 2-[[3,4-bis(chloranyl)-5-methyl-1~{H}-pyrrol-2-yl]carbonylamino]-4-morpholin-4-yl-1,3-benzothiazole-6-carboxylic acid, CALCIUM ION, DNA gyrase subunit B | | Authors: | Durcik, M, Zega, A, Zidar, N, Ilas, J, Tomasic, T, Masic, L.P, Mundy, J.E.A, Stevenson, C.E.M, Burton, N, Lawson, D.M, Maxwell, A, Kikelj, D. | | Deposit date: | 2022-11-12 | | Release date: | 2023-03-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | New Dual Inhibitors of Bacterial Topoisomerases with Broad-Spectrum Antibacterial Activity and In Vivo Efficacy against Vancomycin-Intermediate Staphylococcus aureus .
J.Med.Chem., 66, 2023
|
|
1AB4
 
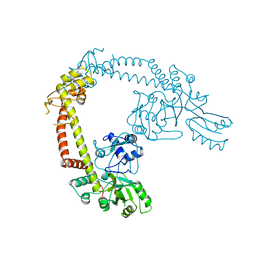 | |
2Y3P
 
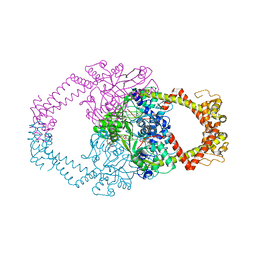 | | Crystal structure of N-terminal domain of GyrA with the antibiotic simocyclinone D8 | | Descriptor: | DNA GYRASE SUBUNIT A, MAGNESIUM ION, SIMOCYCLINONE D8 | | Authors: | Edwards, M.J, Flatman, R.H, Mitchenall, L.A, Stevenson, C.E.M, Le, T.B.K, Clarke, T.A, McKay, A.R, Fiedler, H.-P, Buttner, M.J, Lawson, D.M, Maxwell, A. | | Deposit date: | 2010-12-22 | | Release date: | 2010-12-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | A Crystal Structure of the Bifunctional Antibiotic Simocyclinone D8, Bound to DNA Gyrase.
Science, 326, 2009
|
|
6ZT4
 
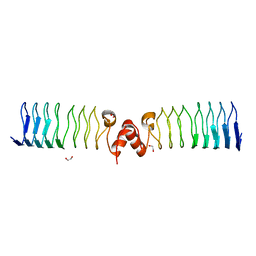 | | Pentapeptide repeat protein MfpA from Mycobacterium smegmatis | | Descriptor: | 1,2-ETHANEDIOL, Pentapeptide repeat protein MfpA | | Authors: | Feng, L, Mundy, J.E.A, Stevenson, C.E.M, Mitchenall, L.A, Lawson, D.M, Mi, K, Maxwell, A. | | Deposit date: | 2020-07-17 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | The pentapeptide-repeat protein, MfpA, interacts with mycobacterial DNA gyrase as a DNA T-segment mimic.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6ZT5
 
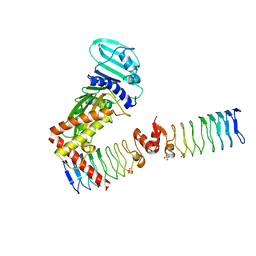 | | Complex between a homodimer of Mycobacterium smegmatis MfpA and a single copy of the N-terminal 47 kDa fragment of the Mycobacterium smegmatis DNA Gyrase B subunit | | Descriptor: | DNA gyrase subunit B, Pentapeptide repeat protein MfpA, SULFATE ION | | Authors: | Feng, L, Mundy, J.E.A, Stevenson, C.E.M, Mitchenall, L.A, Lawson, D.M, Mi, K, Maxwell, A. | | Deposit date: | 2020-07-17 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The pentapeptide-repeat protein, MfpA, interacts with mycobacterial DNA gyrase as a DNA T-segment mimic.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6ZT3
 
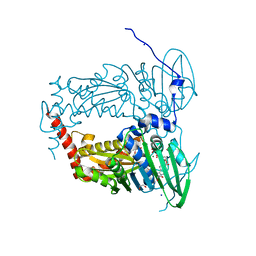 | | N-terminal 47 kDa fragment of the Mycobacterium smegmatis DNA Gyrase B subunit complexed with ADPNP | | Descriptor: | 1,2-ETHANEDIOL, DNA gyrase subunit B, MAGNESIUM ION, ... | | Authors: | Feng, L, Mundy, J.E.A, Stevenson, C.E.M, Mitchenall, L.A, Lawson, D.M, Mi, K, Maxwell, A. | | Deposit date: | 2020-07-17 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | The pentapeptide-repeat protein, MfpA, interacts with mycobacterial DNA gyrase as a DNA T-segment mimic.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
5NJF
 
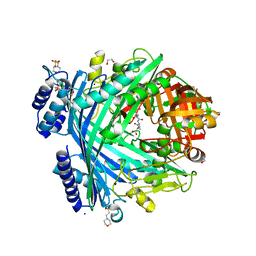 | | E. coli Microcin-processing metalloprotease TldD/E (TldD H262A mutant) with pentapeptide bound | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ALA-ALA-ALA-ALA-ALA, ... | | Authors: | Ghilarov, D, Serebryakova, M, Stevenson, C.E.M, Hearnshaw, S.J, Volkov, D, Maxwell, A, Lawson, D.M, Severinov, K. | | Deposit date: | 2017-03-28 | | Release date: | 2017-10-04 | | Last modified: | 2025-10-01 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | The Origins of Specificity in the Microcin-Processing Protease TldD/E.
Structure, 25, 2017
|
|
5NJB
 
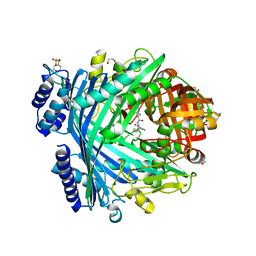 | | E. coli Microcin-processing metalloprotease TldD/E with actinonin bound | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ACTINONIN, ... | | Authors: | Ghilarov, D, Serebryakova, M, Stevenson, C.E.M, Hearnshaw, S.J, Volkov, D, Maxwell, A, Lawson, D.M, Severinov, K. | | Deposit date: | 2017-03-28 | | Release date: | 2017-10-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Origins of Specificity in the Microcin-Processing Protease TldD/E.
Structure, 25, 2017
|
|
4XTJ
 
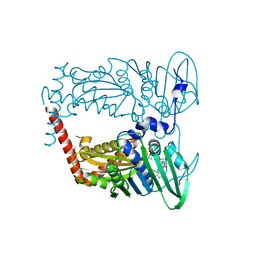 | | N-terminal 43 kDa fragment of the E. coli DNA gyrase B subunit grown from 100 mM KCl plus 100 mM NaCl condition | | Descriptor: | CHLORIDE ION, DNA gyrase subunit B, MAGNESIUM ION, ... | | Authors: | Hearnshaw, S.J, Chung, T.T, Stevenson, C.E.M, Maxwell, A, Lawson, D.M. | | Deposit date: | 2015-01-23 | | Release date: | 2015-04-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | The role of monovalent cations in the ATPase reaction of DNA gyrase
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4WUC
 
 | | N-terminal 43 kDa fragment of the E. coli DNA gyrase B subunit grown from 100 mM NaCl condition | | Descriptor: | CHLORIDE ION, DNA gyrase subunit B, MAGNESIUM ION, ... | | Authors: | Hearnshaw, S.J, Chung, T.T, Stevenson, C.E.M, Maxwell, A, Lawson, D.M. | | Deposit date: | 2014-10-31 | | Release date: | 2015-04-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The role of monovalent cations in the ATPase reaction of DNA gyrase
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4WUD
 
 | | N-terminal 43 kDa fragment of the E. coli DNA gyrase B subunit grown from no salt condition | | Descriptor: | CHLORIDE ION, DNA gyrase subunit B, MAGNESIUM ION, ... | | Authors: | Hearnshaw, S.J, Chung, T.T, Stevenson, C.E.M, Maxwell, A, Lawson, D.M. | | Deposit date: | 2014-10-31 | | Release date: | 2015-04-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The role of monovalent cations in the ATPase reaction of DNA gyrase
Acta Crystallogr.,Sect.D, 71, 2015
|
|
6Z1A
 
 | | Ternary complex of Staphylococcus aureus DNA gyrase with AMK12 and DNA | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Kolaric, A, Germe, T, Hrast, M, Stevenson, C.E.M, Lawson, D.M, Burton, N, Voros, J, Maxwell, A, Minovski, N, Anderluh, M. | | Deposit date: | 2020-05-13 | | Release date: | 2020-11-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Potent DNA gyrase inhibitors bind asymmetrically to their target using symmetrical bifurcated halogen bonds.
Nat Commun, 12, 2021
|
|
6Y8O
 
 | | Mycobacterium smegmatis GyrB 22kDa ATPase sub-domain in complex with novobiocin | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, DNA gyrase subunit B, ... | | Authors: | Henderson, S.R, Stevenson, C.E.M, Malone, B, Zholnerovych, Y, Mitchenall, L.A, Pichowicz, M, McGarry, D.H, Cooper, I.R, Charrier, C, Salisbury, A, Lawson, D.M, Maxwell, A. | | Deposit date: | 2020-03-05 | | Release date: | 2020-08-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and mechanistic analysis of ATPase inhibitors targeting mycobacterial DNA gyrase.
J.Antimicrob.Chemother., 75, 2020
|
|
6Y8L
 
 | | Mycobacterium thermoresistibile GyrB21 in complex with novobiocin | | Descriptor: | 1,2-ETHANEDIOL, DNA gyrase subunit B, NOVOBIOCIN, ... | | Authors: | Henderson, S.R, Stevenson, C.E.M, Malone, B, Zholnerovych, Y, Mitchenall, L.A, Pichowicz, M, McGarry, D.H, Cooper, I.R, Charrier, C, Salisbury, A, Lawson, D.M, Maxwell, A. | | Deposit date: | 2020-03-05 | | Release date: | 2020-08-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural and mechanistic analysis of ATPase inhibitors targeting mycobacterial DNA gyrase.
J.Antimicrob.Chemother., 75, 2020
|
|
6Y8N
 
 | | Mycobacterium thermoresistibile GyrB21 in complex with Redx03863 | | Descriptor: | 1,2-ETHANEDIOL, 4-[(1~{S},5~{R})-6-azanyl-3-azabicyclo[3.1.0]hexan-3-yl]-6-fluoranyl-~{N}-methyl-2-(2-methylpyrimidin-5-yl)oxy-9~{H}-pyrimido[4,5-b]indol-8-amine, DNA gyrase subunit B, ... | | Authors: | Henderson, S.R, Stevenson, C.E.M, Malone, B, Zholnerovych, Y, Mitchenall, L.A, Pichowicz, M, McGarry, D.H, Cooper, I.R, Charrier, C, Salisbury, A, Lawson, D.M, Maxwell, A. | | Deposit date: | 2020-03-05 | | Release date: | 2020-08-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural and mechanistic analysis of ATPase inhibitors targeting mycobacterial DNA gyrase.
J.Antimicrob.Chemother., 75, 2020
|
|
2Y2Z
 
 | | ligand-free form of TetR-like repressor SimR | | Descriptor: | PUTATIVE REPRESSOR SIMREG2 | | Authors: | Le, T.B.K, Stevenson, C.E.M, Fiedler, H.-P, Maxwell, A, Lawson, D.M, Buttner, M.J. | | Deposit date: | 2010-12-17 | | Release date: | 2011-03-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structures of the Tetr-Like Simocyclinone Efflux Pump Repressor, Simr, and the Mechanism of Ligand-Mediated Derepression.
J.Mol.Biol., 408, 2011
|
|
4CKK
 
 | | Apo structure of 55 kDa N-terminal domain of E. coli DNA gyrase A subunit | | Descriptor: | DNA GYRASE SUBUNIT A | | Authors: | Hearnshaw, S.J, Edwards, M.J, Stevenson, C.E.M, Lawson, D.M, Maxwell, A. | | Deposit date: | 2014-01-07 | | Release date: | 2014-03-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A New Crystal Structure of the Bifunctional Antibiotic Simocyclinone D8 Bound to DNA Gyrase Gives Fresh Insight Into the Mechanism of Inhibition.
J.Mol.Biol., 426, 2014
|
|
4CKL
 
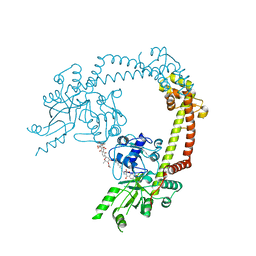 | | Structure of 55 kDa N-terminal domain of E. coli DNA gyrase A subunit with simocyclinone D8 bound | | Descriptor: | DNA GYRASE SUBUNIT A, SIMOCYCLINONE D8 | | Authors: | Hearnshaw, S.J, Edwards, M.J, Stevenson, C.E.M, Lawson, D.M, Maxwell, A. | | Deposit date: | 2014-01-07 | | Release date: | 2014-03-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | A New Crystal Structure of the Bifunctional Antibiotic Simocyclinone D8 Bound to DNA Gyrase Gives Fresh Insight Into the Mechanism of Inhibition.
J.Mol.Biol., 426, 2014
|
|
2Y30
 
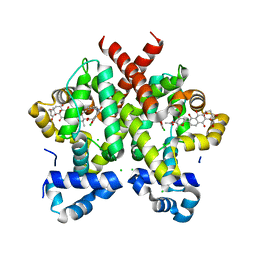 | | Simocyclinone D8 bound form of TetR-like repressor SimR | | Descriptor: | CHLORIDE ION, PUTATIVE REPRESSOR SIMREG2, SIMOCYCLINONE D8 | | Authors: | Le, T.B.K, Stevenson, C.E.M, Fiedler, H.-P, Maxwell, A, Lawson, D.M, Buttner, M.J. | | Deposit date: | 2010-12-17 | | Release date: | 2011-03-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of the Tetr-Like Simocyclinone Efflux Pump Repressor, Simr, and the Mechanism of Ligand-Mediated Derepression.
J.Mol.Biol., 408, 2011
|
|
2Y31
 
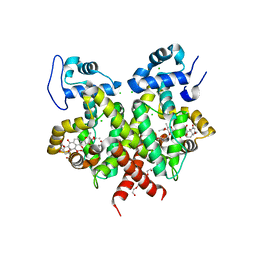 | | Simocyclinone C4 bound form of TetR-like repressor SimR | | Descriptor: | CALCIUM ION, CHLORIDE ION, PENTAETHYLENE GLYCOL, ... | | Authors: | Le, T.B.K, Stevenson, C.E.M, Fiedler, H.-P, Maxwell, A, Lawson, D.M, Buttner, M.J. | | Deposit date: | 2010-12-17 | | Release date: | 2011-03-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of the Tetr-Like Simocyclinone Efflux Pump Repressor, Simr, and the Mechanism of Ligand-Mediated Derepression.
J.Mol.Biol., 408, 2011
|
|
6F8J
 
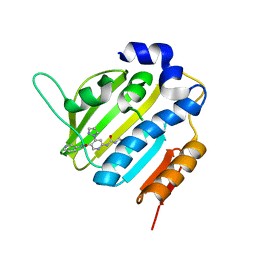 | | Crystal Structure of E. coli GyraseB 24kDa in complex with 6-[(ethylcarbamoyl)amino]-4-(1H-pyrazol-1-yl)-N-(pyridin-3-yl)pyridine-3-carboxamide | | Descriptor: | 6-(ethylcarbamoylamino)-4-pyrazol-1-yl-~{N}-pyridin-3-yl-pyridine-3-carboxamide, DNA gyrase subunit B | | Authors: | Narramore, S.K, Stevenson, C.E.M, Lawson, D.M, Maxwell, A, Fishwick, C.W.G. | | Deposit date: | 2017-12-13 | | Release date: | 2019-06-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | New insights into the binding mode of pyridine-3-carboxamide inhibitors of E. coli DNA gyrase.
Bioorg.Med.Chem., 27, 2019
|
|
6F96
 
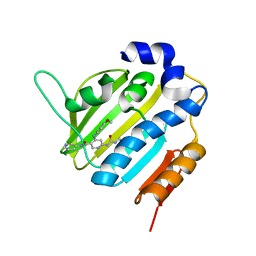 | | Crystal Structure of E. coli GyraseB 24kDa in complex with 6-[(ethylcarbamoyl)amino]-4-[(4-methoxyphenyl)amino]-N-(pyridin-3-yl)pyridine-3-carboxamide | | Descriptor: | 6-(ethylcarbamoylamino)-4-[(4-methoxyphenyl)amino]-~{N}-pyridin-3-yl-pyridine-3-carboxamide, DNA gyrase subunit B | | Authors: | Narramore, S.K, Stevenson, C.E.M, Lawson, D.M, Maxwell, A, Fishwick, C.W.G. | | Deposit date: | 2017-12-14 | | Release date: | 2019-01-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | New insights into the binding mode of pyridine-3-carboxamide inhibitors of E. coli DNA gyrase.
Bioorg.Med.Chem., 27, 2019
|
|
