6LNA
 
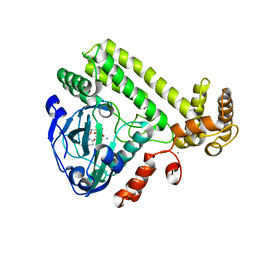 | | YdiU complex with AMPNPP and Mn2+ | | 分子名称: | CALCIUM ION, MANGANESE (II) ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | 著者 | Li, B, Yang, Y, Ma, Y. | | 登録日 | 2019-12-28 | | 公開日 | 2020-12-30 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.701 Å) | | 主引用文献 | The YdiU Domain Modulates Bacterial Stress Signaling through Mn 2+ -Dependent UMPylation.
Cell Rep, 32, 2020
|
|
3Q6S
 
 | |
8SAI
 
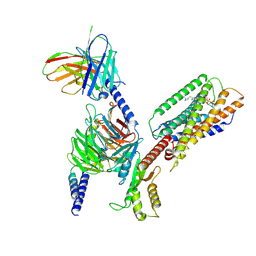 | | Cryo-EM structure of GPR34-Gi complex | | 分子名称: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | 著者 | Yong, X.H, Zhao, C, Yan, W, Shao, Z.H. | | 登録日 | 2023-04-01 | | 公開日 | 2023-10-04 | | 実験手法 | ELECTRON MICROSCOPY (3.27 Å) | | 主引用文献 | Cryo-EM structures of human GPR34 enable the identification of selective antagonists.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
6IZH
 
 | |
8Y3X
 
 | |
7BZV
 
 | |
3N6N
 
 | | crystal structure of EV71 RdRp in complex with Br-UTP | | 分子名称: | 5-bromouridine 5'-(tetrahydrogen triphosphate), NICKEL (II) ION, RNA-dependent RNA polymerase | | 著者 | Wu, Y, Lou, Z.Y, Miao, Y, Yu, Y, Rao, Z.H. | | 登録日 | 2010-05-26 | | 公開日 | 2011-06-15 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Structures of EV71 RNA-dependent RNA polymerase in complex with substrate and analogue provide a drug target against the hand-foot-and-mouth disease pandemic in China.
Protein Cell, 1, 2010
|
|
3N6L
 
 | |
3N6M
 
 | | Crystal structure of EV71 RdRp in complex with GTP | | 分子名称: | GUANOSINE-5'-TRIPHOSPHATE, NICKEL (II) ION, RNA-dependent RNA polymerase | | 著者 | Wu, Y, Lou, Z.Y, Miao, Y, Yu, Y, Rao, Z.H. | | 登録日 | 2010-05-26 | | 公開日 | 2011-06-15 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structures of EV71 RNA-dependent RNA polymerase in complex with substrate and analogue provide a drug target against the hand-foot-and-mouth disease pandemic in China.
Protein Cell, 1, 2010
|
|
7DIY
 
 | | Crystal structure of SARS-CoV-2 nsp10 bound to nsp14-exoribonuclease domain | | 分子名称: | MAGNESIUM ION, ZINC ION, nsp10 protein, ... | | 著者 | Lin, S, Chen, H, Chen, Z.M, Yang, F.L, Ye, F, Zheng, Y, Yang, J, Lin, X, Sun, H.L, Wang, L.L, Wen, A, Cao, Y, Lu, G.W. | | 登録日 | 2020-11-19 | | 公開日 | 2021-05-19 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.693 Å) | | 主引用文献 | Crystal structure of SARS-CoV-2 nsp10 bound to nsp14-ExoN domain reveals an exoribonuclease with both structural and functional integrity.
Nucleic Acids Res., 49, 2021
|
|
8H5T
 
 | |
8H5U
 
 | | Crystal structure of SARS-CoV-2 spike receptor-binding domain in complex with neutralizing nanobody Nb-021 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Nanobody Nb-021, ... | | 著者 | Yang, J, Lin, S, Lu, G.W. | | 登録日 | 2022-10-13 | | 公開日 | 2023-10-18 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.401 Å) | | 主引用文献 | Development of a bispecific nanobody conjugate broadly neutralizes diverse SARS-CoV-2 variants and structural basis for its broad neutralization.
Plos Pathog., 19, 2023
|
|
8IYX
 
 | | Cryo-EM structure of the GPR34 receptor in complex with the antagonist YL-365 | | 分子名称: | 1-[4-(3-chlorophenyl)phenyl]carbonyl-4-[2-(4-phenylmethoxyphenyl)ethanoylamino]piperidine-4-carboxylic acid, Probable G-protein coupled receptor 34,YL-365 | | 著者 | Jia, G.W, Wang, X, Zhang, C.B, Dong, H.H, Su, Z.M. | | 登録日 | 2023-04-06 | | 公開日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (3.34 Å) | | 主引用文献 | Cryo-EM structures of human GPR34 enable the identification of selective antagonists.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
3P8Z
 
 | | Dengue Methyltransferase bound to a SAM-based inhibitor | | 分子名称: | (S)-2-amino-4-(((2S,3S,4R,5R)-5-(6-(3-chlorobenzylamino)-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl)methylthio)butanoic acid, Non-structural protein 5, S-ADENOSYL-L-HOMOCYSTEINE | | 著者 | Noble, C.G, Yap, L.J, Lescar, J. | | 登録日 | 2010-10-15 | | 公開日 | 2010-12-08 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Small molecule inhibitors that selectively block dengue virus methyltransferase
J.Biol.Chem., 286, 2011
|
|
3P97
 
 | |
4L3P
 
 | | Crystal Structure of 2-(1-benzothiophen-7-yl)-4-[1-(piperidin-4-yl)-1H-pyrazol-4-yl]furo[2,3-c]pyridin-7-amine bound to TAK1-TAB1 | | 分子名称: | 2-(1-benzothiophen-7-yl)-4-[1-(piperidin-4-yl)-1H-pyrazol-4-yl]furo[2,3-c]pyridin-7-amine, Mitogen-activated protein kinase kinase kinase 7, TGF-beta-activated kinase 1 and MAP3K7-binding protein 1 chimera | | 著者 | Wang, J, Hornberger, K.R, Crew, A.P, Steinbacher, S, Maskos, K, Moertl, M. | | 登録日 | 2013-06-06 | | 公開日 | 2013-06-19 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.68 Å) | | 主引用文献 | Discovery and optimization of 7-aminofuro[2,3-c]pyridine inhibitors of TAK1.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4L52
 
 | | Crystal Structure of 1-(4-{4-[7-amino-2-(1,2,3-benzothiadiazol-7-yl)furo[2,3-c]pyridin-4-yl]-1H-pyrazol-1-yl}piperidin-1-yl)ethan-1-one bound to TAK1-TAB1 | | 分子名称: | 1-(4-{4-[7-amino-2-(1,2,3-benzothiadiazol-7-yl)furo[2,3-c]pyridin-4-yl]-1H-pyrazol-1-yl}piperidin-1-yl)ethanone, Mitogen-activated protein kinase kinase kinase 7, TGF-beta-activated kinase 1 and MAP3K7-binding protein 1 chimera | | 著者 | Wang, J, Hornberger, K.R, Crew, A.P, Jestel, A, Maskos, K, Moertl, M. | | 登録日 | 2013-06-10 | | 公開日 | 2013-07-03 | | 最終更新日 | 2017-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.54 Å) | | 主引用文献 | Discovery and optimization of 7-aminofuro[2,3-c]pyridine inhibitors of TAK1.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
8GYA
 
 | |
8GY9
 
 | |
8GYB
 
 | |
8GY4
 
 | | Crystal structure of Alongshan virus methyltransferase | | 分子名称: | Methyltransferase | | 著者 | Chen, H, Lin, S, Lu, G.W. | | 登録日 | 2022-09-21 | | 公開日 | 2023-09-27 | | 最終更新日 | 2024-04-10 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structural and functional basis of low-affinity SAM/SAH-binding in the conserved MTase of the multi-segmented Alongshan virus distantly related to canonical unsegmented flaviviruses.
Plos Pathog., 19, 2023
|
|
5ZVD
 
 | |
5ZVG
 
 | |
5I3P
 
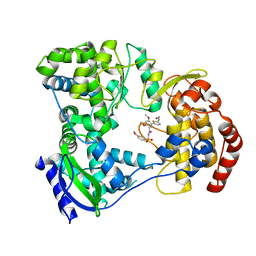 | | DENGUE SEROTYPE 3 RNA-DEPENDENT RNA POLYMERASE BOUND TO COMPOUND 27 | | 分子名称: | 5-[5-(3-hydroxyprop-1-yn-1-yl)thiophen-2-yl]-2,4-dimethoxy-N-[(3-methoxyphenyl)sulfonyl]benzamide, Genome polyprotein, ZINC ION | | 著者 | Noble, C.G. | | 登録日 | 2016-02-10 | | 公開日 | 2016-07-06 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.45 Å) | | 主引用文献 | Potent Allosteric Dengue Virus NS5 Polymerase Inhibitors: Mechanism of Action and Resistance Profiling
Plos Pathog., 12, 2016
|
|
5ZVE
 
 | |
