8JKL
 
 | | IRF4 DNA-binding domain bound to an DNA containing GATA motif | | Descriptor: | GATA-Forward, GATA-Reverse, Interferon regulatory factor 4 | | Authors: | Wang, G, Feng, X, Ding, J. | | Deposit date: | 2023-06-01 | | Release date: | 2023-09-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Molecular basis for the functional roles of the multimorphic T95R mutation of IRF4 causing human autosomal dominant combined immunodeficiency.
Structure, 31, 2023
|
|
8JKQ
 
 | |
8JKN
 
 | |
8JKO
 
 | |
4N3Y
 
 | |
3PPV
 
 | | Crystal structure of an engineered VWF A2 domain (N1493C and C1670S) | | Descriptor: | CALCIUM ION, SULFATE ION, von Willebrand factor | | Authors: | Zhou, M, Dong, X, Zhong, C, Ding, J. | | Deposit date: | 2010-11-25 | | Release date: | 2011-05-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A novel calcium-binding site of von Willebrand factor A2 domain regulates its cleavage by ADAMTS13
Blood, 117, 2011
|
|
3PPX
 
 | | Crystal structure of the N1602A mutant of an engineered VWF A2 domain (N1493C and C1670S) | | Descriptor: | SODIUM ION, von Willebrand factor | | Authors: | Zhou, M, Dong, X, Zhong, C, Ding, J. | | Deposit date: | 2010-11-25 | | Release date: | 2011-05-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | A novel calcium-binding site of von Willebrand factor A2 domain regulates its cleavage by ADAMTS13
Blood, 117, 2011
|
|
3PPW
 
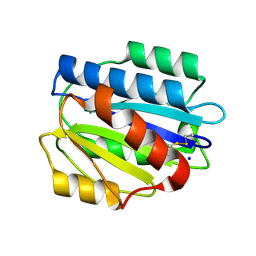 | | Crystal structure of the D1596A mutant of an engineered VWF A2 domain (N1493C and C1670S) | | Descriptor: | SODIUM ION, von Willebrand factor | | Authors: | Zhou, M, Dong, X, Zhong, C, Ding, J. | | Deposit date: | 2010-11-25 | | Release date: | 2011-05-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A novel calcium-binding site of von Willebrand factor A2 domain regulates its cleavage by ADAMTS13
Blood, 117, 2011
|
|
3PPY
 
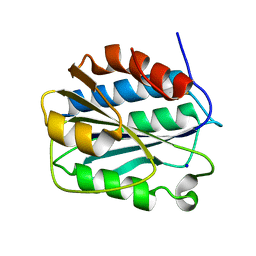 | | Crystal structure of the D1596A/N1602A double mutant of an engineered VWF A2 domain (N1493C and C1670S) | | Descriptor: | SODIUM ION, von Willebrand factor | | Authors: | Zhou, M, Dong, X, Zhong, C, Ding, J. | | Deposit date: | 2010-11-25 | | Release date: | 2011-05-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A novel calcium-binding site of von Willebrand factor A2 domain regulates its cleavage by ADAMTS13
Blood, 117, 2011
|
|
3QAH
 
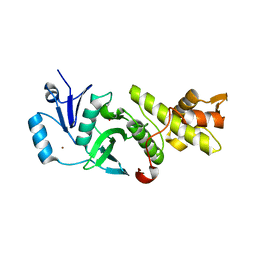 | | Crystal structure of apo-form human MOF catalytic domain | | Descriptor: | Probable histone acetyltransferase MYST1, ZINC ION | | Authors: | Sun, B, Tang, Q, Zhong, C, Ding, J. | | Deposit date: | 2011-01-11 | | Release date: | 2011-07-06 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Regulation of the histone acetyltransferase activity of hMOF via autoacetylation of Lys274
Cell Res., 21, 2011
|
|
3RPN
 
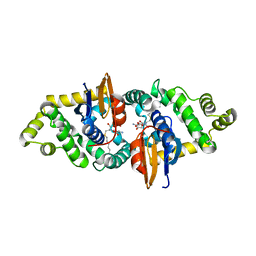 | | Crystal structure of human kappa class glutathione transferase in complex with S-hexylglutathione | | Descriptor: | Glutathione S-transferase kappa 1, S-HEXYLGLUTATHIONE | | Authors: | Wang, B, Peng, Y, Zhang, T, Ding, J. | | Deposit date: | 2011-04-27 | | Release date: | 2011-07-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures and kinetic studies of human Kappa class glutathione transferase provide insights into the catalytic mechanism.
Biochem.J., 439, 2011
|
|
3RIB
 
 | | Human lysine methyltransferase Smyd2 in complex with AdoHcy | | Descriptor: | N-lysine methyltransferase SMYD2, S-ADENOSYL-L-HOMOCYSTEINE, SULFATE ION, ... | | Authors: | Xu, S, Zhang, T, Zhong, C, Ding, J. | | Deposit date: | 2011-04-13 | | Release date: | 2011-07-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Structure of human lysine methyltransferase Smyd2 reveals insights into the substrate divergence in Smyd proteins
J Mol Cell Biol, 3, 2011
|
|
3RPP
 
 | |
7WR4
 
 | | Crystal structure of OspC3-calmodulin-caspase-4 complex | | Descriptor: | Calmodulin-1, Caspase-4, OspC3 | | Authors: | Hou, Y.J, Zeng, H, Shao, F, Ding, J. | | Deposit date: | 2022-01-26 | | Release date: | 2023-01-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural mechanisms of calmodulin activation of Shigella effector OspC3 to ADP-riboxanate caspase-4/11 and block pyroptosis.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7WR1
 
 | | P32 of caspase-4 C258A mutant in complex with OspC3 C-terminal ankyrin-repeat domain | | Descriptor: | Caspase-4, OspC3 | | Authors: | Hou, Y.J, Zeng, H, Shao, F, Ding, J. | | Deposit date: | 2022-01-26 | | Release date: | 2023-01-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Structural mechanisms of calmodulin activation of Shigella effector OspC3 to ADP-riboxanate caspase-4/11 and block pyroptosis.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7WR0
 
 | | P32 of caspase-4 C258A mutant | | Descriptor: | Caspase-4 | | Authors: | Hou, Y.J, Zeng, H, Shao, F, Ding, J. | | Deposit date: | 2022-01-26 | | Release date: | 2023-01-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural mechanisms of calmodulin activation of Shigella effector OspC3 to ADP-riboxanate caspase-4/11 and block pyroptosis.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7WR2
 
 | |
7WR5
 
 | | Crystal structure of OspC3-calmodulin-caspase-4 complex binding with 2'-aF-NAD+ | | Descriptor: | Calmodulin-1, Caspase-4, OspC3, ... | | Authors: | Hou, Y.J, Zeng, H, Shao, F, Ding, J. | | Deposit date: | 2022-01-26 | | Release date: | 2023-01-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural mechanisms of calmodulin activation of Shigella effector OspC3 to ADP-riboxanate caspase-4/11 and block pyroptosis.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7WR3
 
 | | Crystal structure of MBP-fused OspC3 in complex with calmodulin | | Descriptor: | Calmodulin-1, MBP-fused OspC3, NICOTINAMIDE, ... | | Authors: | Hou, Y.J, Zeng, H, Shao, F, Ding, J. | | Deposit date: | 2022-01-26 | | Release date: | 2023-01-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structural mechanisms of calmodulin activation of Shigella effector OspC3 to ADP-riboxanate caspase-4/11 and block pyroptosis.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7WR6
 
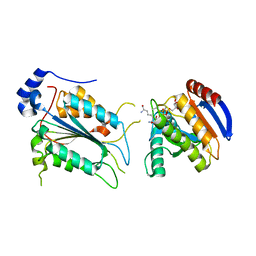 | | Crystal structure of ADP-riboxanated caspase-4 in complex with Af1521 | | Descriptor: | ADP-ribose glycohydrolase AF_1521, Caspase-4, [[(3~{a}~{S},5~{R},6~{R},6~{a}~{R})-2-azanylidene-3-[(4~{R})-4-azanyl-5-oxidanylidene-pentyl]-6-oxidanyl-3~{a},5,6,6~{a}-tetrahydrofuro[2,3-d][1,3]oxazol-5-yl]methoxy-oxidanyl-phosphoryl] [(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl hydrogen phosphate | | Authors: | Hou, Y.J, Zeng, H, Shao, F, Ding, J. | | Deposit date: | 2022-01-26 | | Release date: | 2023-01-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structural mechanisms of calmodulin activation of Shigella effector OspC3 to ADP-riboxanate caspase-4/11 and block pyroptosis.
Nat.Struct.Mol.Biol., 30, 2023
|
|
4N3Z
 
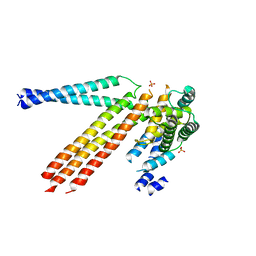 | |
4N3X
 
 | |
7BTA
 
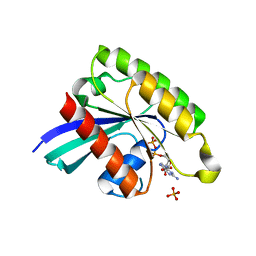 | | Crystal structure of Rheb D60K mutant bound to GDP | | Descriptor: | GTP-binding protein Rheb, GUANOSINE-5'-DIPHOSPHATE, PHOSPHATE ION | | Authors: | Zhang, C, Zhang, T, Ding, J. | | Deposit date: | 2020-03-31 | | Release date: | 2020-06-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Molecular basis for the functions of dominantly active Y35N and inactive D60K Rheb mutants in mTORC1 signaling.
J Mol Cell Biol, 12, 2020
|
|
7BTC
 
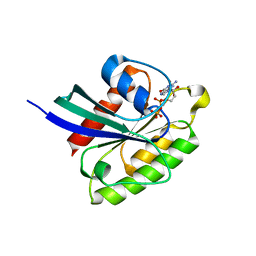 | | Crystal structure of Rheb Y35N mutant bound to GDP | | Descriptor: | GTP-binding protein Rheb, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Zhang, C, Zhang, T, Ding, J. | | Deposit date: | 2020-04-01 | | Release date: | 2020-06-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | Molecular basis for the functions of dominantly active Y35N and inactive D60K Rheb mutants in mTORC1 signaling.
J Mol Cell Biol, 12, 2020
|
|
7BTD
 
 | | Crystal structure of Rheb Y35N mutant bound to GppNHp | | Descriptor: | GTP-binding protein Rheb, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER | | Authors: | Zhang, C, Zhang, T, Ding, J. | | Deposit date: | 2020-04-01 | | Release date: | 2020-06-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular basis for the functions of dominantly active Y35N and inactive D60K Rheb mutants in mTORC1 signaling.
J Mol Cell Biol, 12, 2020
|
|
