4XOI
 
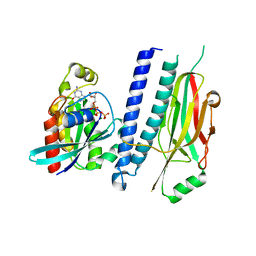 | | Structure of hsAnillin bound with RhoA(Q63L) at 2.1 Angstroms resolution | | Descriptor: | Actin-binding protein anillin, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Sun, L, Guan, R, Lee, I.-J, Liu, Y, Chen, M, Wang, J, Wu, J, Chen, Z. | | Deposit date: | 2015-01-16 | | Release date: | 2015-07-15 | | Last modified: | 2022-03-23 | | Method: | X-RAY DIFFRACTION (2.092 Å) | | Cite: | Mechanistic insights into the anchorage of the contractile ring by anillin and mid1
Dev.Cell, 33, 2015
|
|
4QKF
 
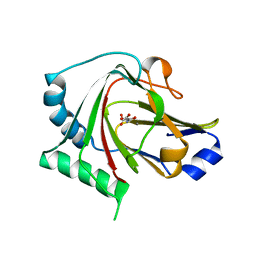 | | Crystal structure of human ALKBH7 in complex with N-oxalylglycine and Mn(II) | | Descriptor: | Alpha-ketoglutarate-dependent dioxygenase alkB homolog 7, mitochondrial, MANGANESE (II) ION, ... | | Authors: | Wang, G, Chen, Z. | | Deposit date: | 2014-06-06 | | Release date: | 2014-08-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | The atomic resolution structure of human AlkB homolog 7 (ALKBH7), a key protein for programmed necrosis and fat metabolism
J.Biol.Chem., 289, 2014
|
|
4QKB
 
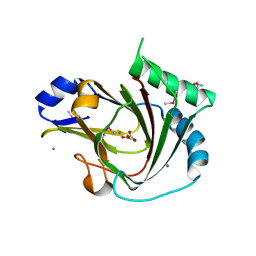 | | Crystal structure of seleno-methionine labelled human ALKBH7 in complex with alpha-ketoglutarate and Mn(II) | | Descriptor: | 2-OXOGLUTARIC ACID, Alpha-ketoglutarate-dependent dioxygenase alkB homolog 7, mitochondrial, ... | | Authors: | Wang, G, He, Q, Chen, Z. | | Deposit date: | 2014-06-05 | | Release date: | 2014-08-20 | | Last modified: | 2015-10-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The atomic resolution structure of human AlkB homolog 7 (ALKBH7), a key protein for programmed necrosis and fat metabolism
J.Biol.Chem., 289, 2014
|
|
4QKD
 
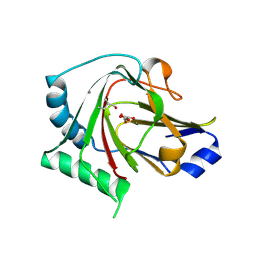 | |
5D0N
 
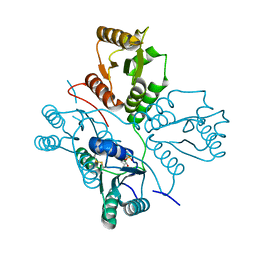 | | Crystal structure of maize PDRP bound with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, MAGNESIUM ION, Pyruvate, ... | | Authors: | Jiang, L, Chen, Z. | | Deposit date: | 2015-08-03 | | Release date: | 2016-02-24 | | Last modified: | 2016-03-09 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural Basis of Reversible Phosphorylation by Maize Pyruvate Orthophosphate Dikinase Regulatory Protein
Plant Physiol., 170, 2016
|
|
5D1F
 
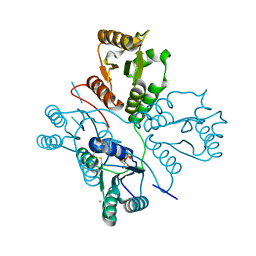 | | Crystal structure of maize PDRP bound with AMP and Hg2+ | | Descriptor: | ADENOSINE MONOPHOSPHATE, MAGNESIUM ION, MERCURY (II) ION, ... | | Authors: | Jiang, L, Chen, Z. | | Deposit date: | 2015-08-04 | | Release date: | 2016-02-24 | | Last modified: | 2020-02-19 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural Basis of Reversible Phosphorylation by Maize Pyruvate Orthophosphate Dikinase Regulatory Protein
Plant Physiol., 170, 2016
|
|
1GG4
 
 | |
5EYI
 
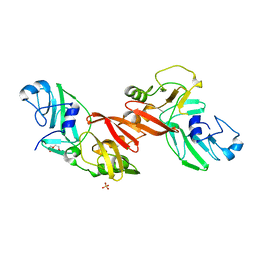 | | Structure of PRRSV apo-NSP11 at 2.16A | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, CHLORIDE ION, Non-structural protein 11, ... | | Authors: | Zhang, M.F, Chen, Z. | | Deposit date: | 2015-11-25 | | Release date: | 2016-10-12 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Structural Biology of the Arterivirus nsp11 Endoribonucleases.
J. Virol., 91, 2017
|
|
3KLX
 
 | |
3KL1
 
 | |
6P9U
 
 | | Crystal structure of human thrombin mutant W215A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Prothrombin, ZINC ION | | Authors: | Pelc, L.A, Koester, S.K, Chen, Z, Di Cera, E. | | Deposit date: | 2019-06-10 | | Release date: | 2019-09-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Residues W215, E217 and E192 control the allosteric E*-E equilibrium of thrombin.
Sci Rep, 9, 2019
|
|
4RN6
 
 | | Structure of prethrombin-2 mutant s195a bound to the active site inhibitor argatroban | | Descriptor: | (2R,4R)-4-methyl-1-(N~2~-{[(3S)-3-methyl-1,2,3,4-tetrahydroquinolin-8-yl]sulfonyl}-L-arginyl)piperidine-2-carboxylic acid, Thrombin heavy chain | | Authors: | Pozzi, N, Chen, Z, Zapata, F, Niu, W, Barranco-Medina, S, Pelc, L.A, Di Cera, E. | | Deposit date: | 2014-10-23 | | Release date: | 2014-11-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Autoactivation of thrombin precursors.
J.Biol.Chem., 288, 2013
|
|
4RKJ
 
 | | Crystal structure of thrombin mutant S195T (free form) | | Descriptor: | GLYCEROL, POTASSIUM ION, Thrombin heavy chain, ... | | Authors: | Pelc, A.L, Chen, Z, Gohara, D.W, Vogt, A.D, Pozzi, N, Di Cera, E. | | Deposit date: | 2014-10-13 | | Release date: | 2015-03-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Why ser and not thr brokers catalysis in the trypsin fold.
Biochemistry, 54, 2015
|
|
4RKO
 
 | | Crystal structure of thrombin mutant S195T bound with PPACK | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, ... | | Authors: | Pelc, A.L, Chen, Z, Gohara, D.W, Vogt, A.D, Pozzi, N, Di Cera, E. | | Deposit date: | 2014-10-13 | | Release date: | 2015-03-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Why ser and not thr brokers catalysis in the trypsin fold.
Biochemistry, 54, 2015
|
|
2PV9
 
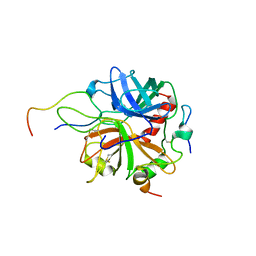 | | Crystal structure of murine thrombin in complex with the extracellular fragment of murine PAR4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Proteinase-activated receptor 4, Thrombin heavy chain, ... | | Authors: | Bah, A, Chen, Z, Bush-Pelc, L.A, Mathews, F.S, Di Cera, E. | | Deposit date: | 2007-05-09 | | Release date: | 2007-07-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal structures of murine thrombin in complex with the extracellular fragments of murine protease-activated receptors PAR3 and PAR4.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2PUX
 
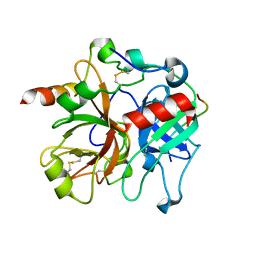 | | Crystal structure of murine thrombin in complex with the extracellular fragment of murine PAR3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Proteinase-activated receptor 3, Thrombin heavy chain, ... | | Authors: | Bah, A, Chen, Z, Bush-Pelc, L.A, Mathews, F.S, Di Cera, E. | | Deposit date: | 2007-05-09 | | Release date: | 2007-07-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of murine thrombin in complex with the extracellular fragments of murine protease-activated receptors PAR3 and PAR4.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
7MIS
 
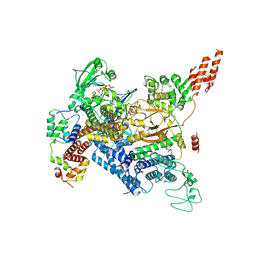 | | Cryo-EM structure of SidJ-SdeC-CaM reaction intermediate complex | | Descriptor: | ADENOSINE MONOPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, ... | | Authors: | Osinski, A, Black, M.H, Pawlowski, K, Chen, Z, Li, Y, Tagliabracci, V.S. | | Deposit date: | 2021-04-17 | | Release date: | 2021-08-18 | | Last modified: | 2021-11-17 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural and mechanistic basis for protein glutamylation by the kinase fold.
Mol.Cell, 81, 2021
|
|
7MIR
 
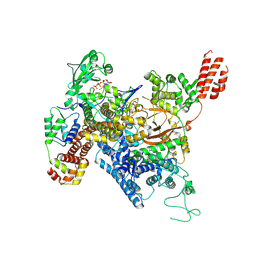 | | Cryo-EM structure of SidJ-SdeA-CaM reaction intermediate complex | | Descriptor: | ADENOSINE MONOPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, ... | | Authors: | Osinski, A, Black, M.H, Pawlowski, K, Chen, Z, Li, Y, Tagliabracci, V.S. | | Deposit date: | 2021-04-17 | | Release date: | 2021-08-18 | | Last modified: | 2021-11-17 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Structural and mechanistic basis for protein glutamylation by the kinase fold.
Mol.Cell, 81, 2021
|
|
3BEI
 
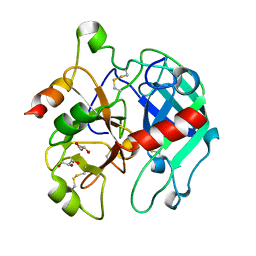 | | Crystal structure of the slow form of thrombin in a self_inhibited conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Prothrombin | | Authors: | Gandhi, P.S, Chen, Z, Mathews, F.S, Di Cera, E. | | Deposit date: | 2007-11-19 | | Release date: | 2007-12-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural identification of the pathway of long-range communication in an allosteric enzyme.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
7NNS
 
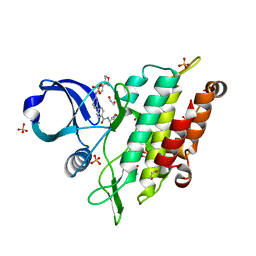 | | Crystal structure of the ACVR1 (ALK2) kinase in complex with the compound Momelotinib | | Descriptor: | 1,2-ETHANEDIOL, Activin receptor type I, Momelotinib, ... | | Authors: | Williams, E, Chen, Z, Burgess-Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A.N. | | Deposit date: | 2021-02-25 | | Release date: | 2021-04-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Crystal structure of the ACVR1 (ALK2) kinase in complex with the compound Momelotinib
To Be Published
|
|
1INP
 
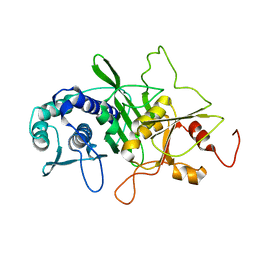 | | CRYSTAL STRUCTURE OF INOSITOL POLYPHOSPHATE 1-PHOSPHATASE AT 2.3 ANGSTROMS RESOLUTION | | Descriptor: | INOSITOL POLYPHOSPHATE 1-PHOSPHATASE, MAGNESIUM ION | | Authors: | York, J.D, Ponder, J.W, Chen, Z, Mathews, F.S, Majerus, P.W. | | Deposit date: | 1994-10-04 | | Release date: | 1995-01-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of inositol polyphosphate 1-phosphatase at 2.3-A resolution.
Biochemistry, 33, 1994
|
|
3EE0
 
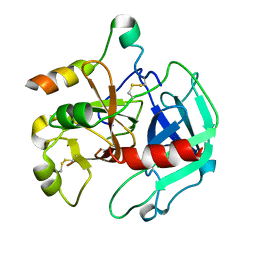 | | Crystal Structure of the W215A/E217A Mutant of Human Thrombin (space group P2(1)2(1)2(1)) | | Descriptor: | Thrombin heavy chain, Thrombin light chain | | Authors: | Gandhi, P.S, Page, M.J, Chen, Z, Bush-Pelc, L, Di Cera, E. | | Deposit date: | 2008-09-03 | | Release date: | 2009-07-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Molecular Basis for the Kinetic Differences of the W215A/E217A Mutant of Human and Murine Thrombin
To be Published
|
|
3EDX
 
 | | Crystal structure of the W215A/E217A mutant of murine thrombin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, SULFATE ION, Thrombin heavy chain, ... | | Authors: | Gandhi, P.S, Page, M.J, Chen, Z, Bush-pelc, L, Di Cera, E. | | Deposit date: | 2008-09-03 | | Release date: | 2009-07-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular Basis for the Kinetic Differences of the W215A/E217A Mutant of Human and Murine Thrombin
To be Published
|
|
7Q5U
 
 | | The tandem SH2 domains of SYK with a bound CD3G diphospho-ITAM peptide | | Descriptor: | 1,2-ETHANEDIOL, 1-ETHOXY-2-(2-ETHOXYETHOXY)ETHANE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Chen, Z, Bountra, C, von Delft, F, Gileadi, O, Brennan, P.E. | | Deposit date: | 2021-11-04 | | Release date: | 2021-11-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The tandem SH2 domains of SYK
To Be Published
|
|
7Q5T
 
 | | The tandem SH2 domains of SYK with a bound FCER1G diphospho-ITAM peptide | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, High affinity immunoglobulin epsilon receptor subunit gamma, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Chen, Z, Bountra, C, von Delft, F, Gileadi, O, Brennan, P.E. | | Deposit date: | 2021-11-04 | | Release date: | 2021-11-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The tandem SH2 domains of SYK
To Be Published
|
|
