6MSR
 
 | | Crystal structure of pRO-2.5 | | Descriptor: | pRO-2.5 | | Authors: | Bick, M.J, Sankaran, B, Boyken, S.E, Baker, D. | | Deposit date: | 2018-10-17 | | Release date: | 2019-05-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | De novo design of tunable, pH-driven conformational changes.
Science, 364, 2019
|
|
3HVV
 
 | |
3HVX
 
 | |
6N9D
 
 | | Complex of tissue inhibitor of metalloproteinases-1 (TIMP-1) mutant (L34G/L133P/L151C/G154A) with matrix metalloproteinase-3 catalytic domain (MMP-3cd) | | Descriptor: | CALCIUM ION, Metalloproteinase inhibitor 1, Stromelysin-1, ... | | Authors: | Raeeszadeh-Sarmazdeh, M, Radisky, E.S, Sankaran, B. | | Deposit date: | 2018-12-03 | | Release date: | 2019-05-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Directed evolution of the metalloproteinase inhibitor TIMP-1 reveals that its N- and C-terminal domains cooperate in matrix metalloproteinase recognition.
J.Biol.Chem., 294, 2019
|
|
6BDL
 
 | |
6BG2
 
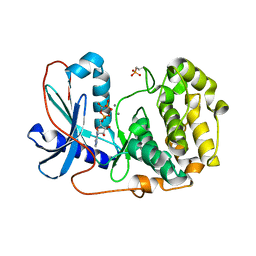 | |
6BX8
 
 | | Human Mesotrypsin (PRSS3) Complexed with Tissue Factor Pathway Inhibitor Variant (TFPI1-KD1-K15R-I17C-I34C) | | Descriptor: | SULFATE ION, Tissue factor pathway inhibitor, Trypsin-3 | | Authors: | Coban, M, Sankaran, B, Cohen, I, Hockla, A, Papo, N, Radisky, E.S. | | Deposit date: | 2017-12-18 | | Release date: | 2019-02-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Disulfide engineering of human Kunitz-type serine protease inhibitors enhances proteolytic stability and target affinity toward mesotrypsin.
J. Biol. Chem., 294, 2019
|
|
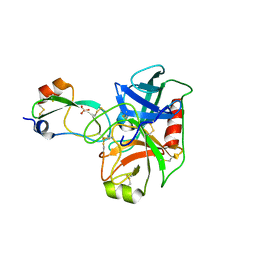
6C0U
 
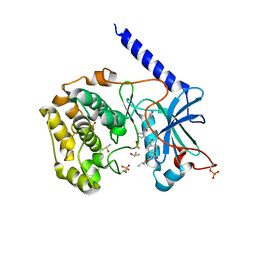 | | Crystal structure of cAMP-dependent protein kinase Calpha subunit bound with N46 | | Descriptor: | DIMETHYL SULFOXIDE, N-[(3R,4R)-4-{[4-(2-fluoro-3-methoxy-6-propoxybenzene-1-carbonyl)benzene-1-carbonyl]amino}pyrrolidin-3-yl]-1H-indazole-5-carboxamide, PHOSPHATE ION, ... | | Authors: | Qin, L, Sankaran, B, Kim, C. | | Deposit date: | 2018-01-02 | | Release date: | 2018-05-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural basis for selective inhibition of human PKG I alpha by the balanol-like compound N46.
J. Biol. Chem., 293, 2018
|
|
6C0T
 
 | | Crystal structure of cGMP-dependent protein kinase Ialpha (PKG Ialpha) catalytic domain bound with N46 | | Descriptor: | DIMETHYL SULFOXIDE, N-[(3R,4R)-4-{[4-(2-fluoro-3-methoxy-6-propoxybenzene-1-carbonyl)benzene-1-carbonyl]amino}pyrrolidin-3-yl]-1H-indazole-5-carboxamide, TRIETHYLENE GLYCOL, ... | | Authors: | Qin, L, Sankaran, B, Kim, C. | | Deposit date: | 2018-01-02 | | Release date: | 2018-05-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structural basis for selective inhibition of human PKG I alpha by the balanol-like compound N46.
J. Biol. Chem., 293, 2018
|
|
6C89
 
 | | NDM-1 Beta-Lactamase Exhibits Differential Active Site Sequence Requirements for the Hydrolysis of Penicillin versus Carbapenem Antibiotics | | Descriptor: | 1,2-ETHANEDIOL, Beta-lactamase, CHLORIDE ION, ... | | Authors: | Palzkill, T, Sun, Z, Sankaran, B. | | Deposit date: | 2018-01-24 | | Release date: | 2018-12-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.75006151 Å) | | Cite: | Differential active site requirements for NDM-1 beta-lactamase hydrolysis of carbapenem versus penicillin and cephalosporin antibiotics.
Nat Commun, 9, 2018
|
|
6CWV
 
 | | Protein Tyrosine Phosphatase 1B A122S mutant | | Descriptor: | MAGNESIUM ION, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Hjortness, M, Zwart, P, Sankaran, B, Fox, J.M. | | Deposit date: | 2018-03-31 | | Release date: | 2018-10-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.98002291 Å) | | Cite: | Evolutionarily Conserved Allosteric Communication in Protein Tyrosine Phosphatases.
Biochemistry, 57, 2018
|
|
6CWU
 
 | | Protein Tyrosine Phosphatase 1B F135Y mutant | | Descriptor: | MAGNESIUM ION, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Hjortness, M, Zwart, P, Sankaran, B, Fox, J.M. | | Deposit date: | 2018-03-31 | | Release date: | 2018-10-31 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Evolutionarily Conserved Allosteric Communication in Protein Tyrosine Phosphatases.
Biochemistry, 57, 2018
|
|
5TWD
 
 | | CTX-M-14 P167S apoenzyme | | Descriptor: | Beta-lactamase | | Authors: | Patel, M, Stojanoski, V, Sankaran, B, Prasad, B.V.V, Palzkill, T. | | Deposit date: | 2016-11-12 | | Release date: | 2017-06-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Drug-Resistant Variant P167S Expands the Substrate Profile of CTX-M beta-Lactamases for Oxyimino-Cephalosporin Antibiotics by Enlarging the Active Site upon Acylation.
Biochemistry, 56, 2017
|
|
5TZO
 
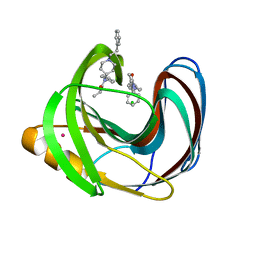 | | Computationally Designed Fentanyl Binder - Fen49*-Complex | | Descriptor: | CHLORIDE ION, Endo-1,4-beta-xylanase A, N-phenyl-N-[1-(2-phenylethyl)piperidin-4-yl]propanamide, ... | | Authors: | Bick, M.J, Greisen, P.J, Morey, K.J, Antunes, M.S, La, D, Sankaran, B, Reymond, L, Johnsson, K, Medford, J.I, Baker, D. | | Deposit date: | 2016-11-22 | | Release date: | 2017-10-04 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Computational design of environmental sensors for the potent opioid fentanyl.
Elife, 6, 2017
|
|
5TWE
 
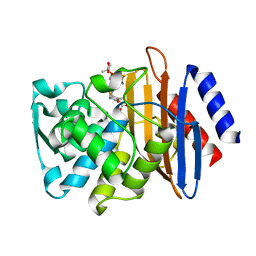 | | CTX-M-14 P167S:S70G mutant enzyme crystallized with ceftazidime | | Descriptor: | ACYLATED CEFTAZIDIME, Beta-lactamase | | Authors: | Patel, M, Stojanoski, V, Sankaran, B, Prasad, B.V.V, Palzkill, T. | | Deposit date: | 2016-11-12 | | Release date: | 2017-06-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Drug-Resistant Variant P167S Expands the Substrate Profile of CTX-M beta-Lactamases for Oxyimino-Cephalosporin Antibiotics by Enlarging the Active Site upon Acylation.
Biochemistry, 56, 2017
|
|
5TW6
 
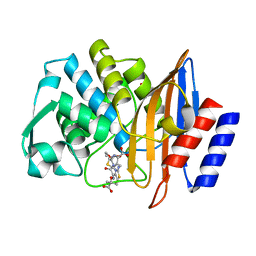 | | CTX-M-14 P167S:E166A mutant with acylated ceftazidime molecule | | Descriptor: | 1,2-ETHANEDIOL, ACYLATED CEFTAZIDIME, Beta-lactamase | | Authors: | Patel, M, Stojanoski, V, Sankaran, B, Prasad, B.V.V, Palzkill, T. | | Deposit date: | 2016-11-11 | | Release date: | 2017-06-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Drug-Resistant Variant P167S Expands the Substrate Profile of CTX-M beta-Lactamases for Oxyimino-Cephalosporin Antibiotics by Enlarging the Active Site upon Acylation.
Biochemistry, 56, 2017
|
|
5TVV
 
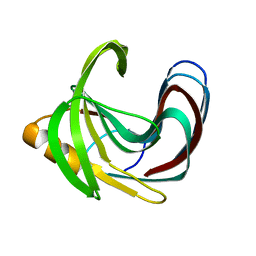 | | Computationally Designed Fentanyl Binder - Fen49* Apo | | Descriptor: | Endo-1,4-beta-xylanase A, POTASSIUM ION | | Authors: | Bick, M.J, Greisen, P.J, Morey, K.J, Antunes, A.S, La, D, Sankaran, B, Reymond, L, Johnsson, K, Medford, J.I, Baker, D. | | Deposit date: | 2016-11-10 | | Release date: | 2017-10-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Computational design of environmental sensors for the potent opioid fentanyl.
Elife, 6, 2017
|
|
5U53
 
 | | CTX-M-14 E166A with acylated ceftazidime molecule | | Descriptor: | ACYLATED CEFTAZIDIME, Beta-lactamase, NITRATE ION | | Authors: | Patel, M, Stojanoski, V, Sankaran, B, Prasad, B.V.V, Palzkill, T. | | Deposit date: | 2016-12-06 | | Release date: | 2017-06-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The Drug-Resistant Variant P167S Expands the Substrate Profile of CTX-M beta-Lactamases for Oxyimino-Cephalosporin Antibiotics by Enlarging the Active Site upon Acylation.
Biochemistry, 56, 2017
|
|
5V0F
 
 | | Crystal structure of C-As lyase with mutation K105A and substrate Roxarsone | | Descriptor: | 4-arsanyl-2-nitrophenol, CHLORIDE ION, FE (III) ION, ... | | Authors: | Venkadesh, S, Yoshinaga, M, Sankaran, B, Kandavelu, P, Rosen, B.P. | | Deposit date: | 2017-02-28 | | Release date: | 2018-03-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Crystal structure of C-As lyase with mutation K105A and substrate Roxarsone.
To Be Published
|
|
5TVY
 
 | | Computationally Designed Fentanyl Binder - Fen49 | | Descriptor: | 3,6,9,12,15,18,21,24,27-NONAOXANONACOSANE-1,29-DIOL, Endo-1,4-beta-xylanase A | | Authors: | Bick, M.J, Greisen, P.J, Morey, K.J, Antunes, M.S, La, D, Sankaran, B, Reymond, L, Johnsson, K, Medford, J.I, Baker, D. | | Deposit date: | 2016-11-10 | | Release date: | 2017-10-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Computational design of environmental sensors for the potent opioid fentanyl.
Elife, 6, 2017
|
|
4S2L
 
 | | Crystal Structure of OXA-163 beta-lactamase | | Descriptor: | Beta-lactamase, SODIUM ION | | Authors: | Stojanoski, V, Liya, H, Palzkill, T.G, Prasad, B, Sankaran, B. | | Deposit date: | 2015-01-21 | | Release date: | 2015-07-22 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Structural Basis for Different Substrate Profiles of Two Closely Related Class D beta-Lactamases and Their Inhibition by Halogens.
Biochemistry, 54, 2015
|
|
6U7V
 
 | | xRRM structure of spPof8 | | Descriptor: | NITRATE ION, Protein pof8 | | Authors: | Kim, J.-K, Hu, X, Yu, C, Jun, H.-I, Liu, J, Sankaran, B, Huang, L, Qiao, F. | | Deposit date: | 2019-09-03 | | Release date: | 2020-09-09 | | Last modified: | 2021-03-24 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Quality-Control Mechanism for Telomerase RNA Folding in the Cell.
Cell Rep, 33, 2020
|
|
6V8V
 
 | | Crystal structure of CTX-M-14 E166A/P167S/D240G beta-lactamase in complex with ceftazidime-2 | | Descriptor: | ACYLATED CEFTAZIDIME, Beta-lactamase | | Authors: | Brown, C.A, Hu, L, Sankaran, B, Prasad, B.V.V, Palzkill, T.G. | | Deposit date: | 2019-12-12 | | Release date: | 2020-04-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Antagonism between substitutions in beta-lactamase explains a path not taken in the evolution of bacterial drug resistance.
J.Biol.Chem., 295, 2020
|
|
6V83
 
 | | Crystal structure of CTX-M-14 E166A/P167S/D240G beta-lactamase in complex with ceftazidime-1 | | Descriptor: | ACYLATED CEFTAZIDIME, Beta-lactamase | | Authors: | Brown, C.A, Hu, L, Sankaran, B, Prasad, B.V.V, Palzkill, T.G. | | Deposit date: | 2019-12-10 | | Release date: | 2020-04-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Antagonism between substitutions in beta-lactamase explains a path not taken in the evolution of bacterial drug resistance.
J.Biol.Chem., 295, 2020
|
|
8ERW
 
 | | Precisely patterned nanofibers made from extendable protein multiplexes | | Descriptor: | C2HR4_8r | | Authors: | Bick, M.J, Parmeggiani, F, Bethel, N.P, Sankaran, B, Baker, D. | | Deposit date: | 2022-10-12 | | Release date: | 2023-08-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Precisely patterned nanofibres made from extendable protein multiplexes.
Nat.Chem., 15, 2023
|
|
