3KXF
 
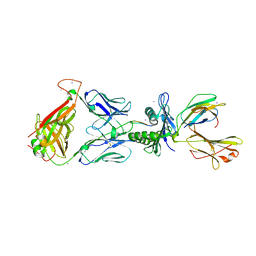 | | Crystal Structure of SB27 TCR in complex with the 'restriction triad' mutant HLA-B*3508-13mer | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, B-35 alpha chain, ... | | Authors: | Archbold, J.K, Tynan, F.E, Gras, S, Rossjohn, J. | | Deposit date: | 2009-12-03 | | Release date: | 2010-06-09 | | Last modified: | 2014-02-26 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Hard wiring of T cell receptor specificity for the major histocompatibility complex is underpinned by TCR adaptability
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
7F45
 
 | | Structure of an Anti-CRISPR protein | | Descriptor: | AcrIF5 | | Authors: | Feng, Y. | | Deposit date: | 2021-06-17 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.52 Å) | | Cite: | AcrIF5 specifically targets DNA-bound CRISPR-Cas surveillance complex for inhibition.
Nat.Chem.Biol., 18, 2022
|
|
7EQG
 
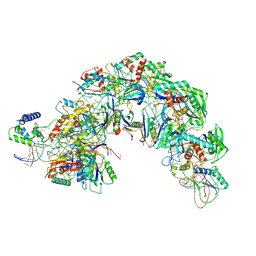 | | Structure of Csy-AcrIF5 | | Descriptor: | AcrIF5, CRISPR type I-F/YPEST-associated protein Csy2, CRISPR-associated protein Csy3, ... | | Authors: | Zhang, L, Feng, Y. | | Deposit date: | 2021-05-01 | | Release date: | 2022-03-09 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | AcrIF5 specifically targets DNA-bound CRISPR-Cas surveillance complex for inhibition.
Nat.Chem.Biol., 18, 2022
|
|
7F3M
 
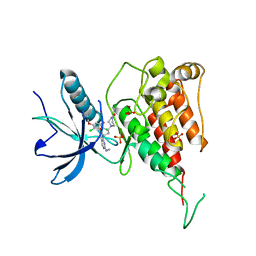 | | Crystal structure of FGFR4 kinase domain with PRN1371 | | Descriptor: | 6-[2,6-bis(chloranyl)-3,5-dimethoxy-phenyl]-2-(methylamino)-8-[3-(4-prop-2-enoylpiperazin-1-yl)propyl]pyrido[2,3-d]pyrimidin-7-one, Fibroblast growth factor receptor 4, SULFATE ION | | Authors: | Chen, X.J, Qu, L.Z, Dai, S.Y, Wei, H.D, Chen, Y.H. | | Deposit date: | 2021-06-16 | | Release date: | 2022-01-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.289 Å) | | Cite: | Structural insights into the potency and selectivity of covalent pan-FGFR inhibitors
Commun Chem, 5, 2022
|
|
6K1P
 
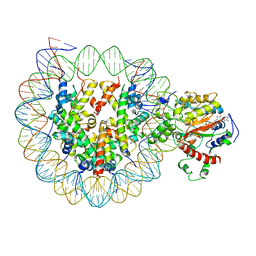 | | The complex of ISWI-nucleosome in the ADP.BeF-bound state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, DNA (167-MER), ... | | Authors: | Yan, L.J, Wu, H, Li, X.M, Gao, N, Chen, Z.C. | | Deposit date: | 2019-05-10 | | Release date: | 2019-05-29 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.87 Å) | | Cite: | Structures of the ISWI-nucleosome complex reveal a conserved mechanism of chromatin remodeling.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6JYL
 
 | | The crosslinked complex of ISWI-nucleosome in the ADP.BeF-bound state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, DNA (167-MER), ... | | Authors: | Yan, L.J, Wu, H, Li, X.M, Gao, N, Chen, Z.C. | | Deposit date: | 2019-04-26 | | Release date: | 2019-05-29 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | Structures of the ISWI-nucleosome complex reveal a conserved mechanism of chromatin remodeling.
Nat.Struct.Mol.Biol., 26, 2019
|
|
3MGT
 
 | | Crystal structure of a H5-specific CTL epitope variant derived from H5N1 influenza virus in complex with HLA-A*0201 | | Descriptor: | 10-meric peptide from Hemagglutinin, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | Authors: | Sun, Y, Liu, J, Yang, M, Gao, F, Zhou, J, Kitamura, Y. | | Deposit date: | 2010-04-07 | | Release date: | 2010-05-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.197 Å) | | Cite: | Identification and structural definition of H5-specific CTL epitopes restricted by HLA-A*0201 derived from the H5N1 subtype of influenza A viruses
J.Gen.Virol., 91, 2010
|
|
7ENN
 
 | | The structure of ALC1 bound to the nucleosome | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Chromodomain-helicase-DNA-binding protein 1-like, ... | | Authors: | Chen, Z.C, Chen, K.J, Wang, L. | | Deposit date: | 2021-04-18 | | Release date: | 2021-07-14 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis of ALC1/CHD1L autoinhibition and the mechanism of activation by the nucleosome.
Nat Commun, 12, 2021
|
|
5U17
 
 | |
5U2V
 
 | |
5U16
 
 | | Structure of human MR1-2-OH-1-NA in complex with human MAIT A-F7 TCR | | Descriptor: | 2-hydroxynaphthalene-1-carbaldehyde, Beta-2-microglobulin, CHLORIDE ION, ... | | Authors: | Keller, A.N, Birkinshaw, R.W, Rossjohn, J. | | Deposit date: | 2016-11-27 | | Release date: | 2017-02-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Drugs and drug-like molecules can modulate the function of mucosal-associated invariant T cells.
Nat. Immunol., 18, 2017
|
|
5W0E
 
 | | CREBBP bromodomain in complex with Cpd19 (3-(7-(difluoromethyl)-6-(1-methyl-1H-pyrazol-4-yl)-3,4-dihydroquinolin-1(2H)-yl)-N-methyl-1-(tetrahydro-2H-pyran-4-yl)-1,4,6,7-tetrahydro-5H-pyrazolo[4,3-c]pyridine-5-carboxamide) | | Descriptor: | 3-[7-(difluoromethyl)-6-(1-methyl-1H-pyrazol-4-yl)-3,4-dihydroquinolin-1(2H)-yl]-N-methyl-1-(oxan-4-yl)-1,4,6,7-tetrahydro-5H-pyrazolo[4,3-c]pyridine-5-carboxamide, CREB-binding protein | | Authors: | Murray, J.M. | | Deposit date: | 2017-05-30 | | Release date: | 2018-02-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | A Unique Approach to Design Potent and Selective Cyclic Adenosine Monophosphate Response Element Binding Protein, Binding Protein (CBP) Inhibitors.
J. Med. Chem., 60, 2017
|
|
7FIA
 
 | | Structure of AcrIF23 | | Descriptor: | AcrIF23 | | Authors: | Ren, J, Yue, F. | | Deposit date: | 2021-07-30 | | Release date: | 2022-07-27 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Structural and mechanistic insights into the inhibition of type I-F CRISPR-Cas system by anti-CRISPR protein AcrIF23.
J.Biol.Chem., 298, 2022
|
|
7F61
 
 | |
7FAS
 
 | | VAR2CSA 3D7 ectodomain core region | | Descriptor: | Erythrocyte membrane protein 1, PfEMP1 | | Authors: | Wang, L, Zhaoning, W. | | Deposit date: | 2021-07-07 | | Release date: | 2021-11-17 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | The molecular mechanism of cytoadherence to placental or tumor cells through VAR2CSA from Plasmodium falciparum.
Cell Discov, 7, 2021
|
|
1BG0
 
 | | TRANSITION STATE STRUCTURE OF ARGININE KINASE | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ARGININE KINASE, D-ARGININE, ... | | Authors: | Zhou, G, Somasundaram, T, Blanc, E, Parthasarathy, G, Ellington, W.R, Chapman, M.S. | | Deposit date: | 1998-06-03 | | Release date: | 1998-10-14 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Transition state structure of arginine kinase: implications for catalysis of bimolecular reactions.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
6K81
 
 | | Crystal structure of human VASH1-SVBP complex | | Descriptor: | Small vasohibin-binding protein, Tubulinyl-Tyr carboxypeptidase 1 | | Authors: | Liu, X, Wang, H, Zhang, Y, Feng, Y. | | Deposit date: | 2019-06-11 | | Release date: | 2020-02-19 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structural insights into tubulin detyrosination by vasohibins-SVBP complex.
Cell Discov, 5, 2019
|
|
7C2J
 
 | | Crystal structure of nsp16-nsp10 heterodimer from SARS-CoV-2 in complex with SAM (without additional SAM during crystallization) | | Descriptor: | 2'-O-methyltransferase, Non-structural protein 10, S-ADENOSYLMETHIONINE, ... | | Authors: | Lin, S, Chen, H, Ye, F, Chen, Z.M, Yang, F.L, Zheng, Y, Cao, Y, Qiao, J.X, Yang, S.Y, Lu, G.W. | | Deposit date: | 2020-05-07 | | Release date: | 2020-05-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.799 Å) | | Cite: | Crystal structure of SARS-CoV-2 nsp10/nsp16 2'-O-methylase and its implication on antiviral drug design.
Signal Transduct Target Ther, 5, 2020
|
|
7C2I
 
 | | Crystal structure of nsp16-nsp10 heterodimer from SARS-CoV-2 in complex with SAM (with additional SAM during crystallization) | | Descriptor: | 2'-O-methyltransferase, Non-structural protein 10, S-ADENOSYLMETHIONINE, ... | | Authors: | Lin, S, Chen, H, Ye, F, Chen, Z.M, Yang, F.L, Zheng, Y, Cao, Y, Qiao, J.X, Yang, S.Y, Lu, G.W. | | Deposit date: | 2020-05-07 | | Release date: | 2020-05-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of SARS-CoV-2 nsp10/nsp16 2'-O-methylase and its implication on antiviral drug design.
Signal Transduct Target Ther, 5, 2020
|
|
5JFR
 
 | | Potent, Reversible MetAP2 Inhibitors via Fragment Based Drug Discovery | | Descriptor: | 1,2-ETHANEDIOL, 7-fluoro-4-(5-methyl-3H-imidazo[4,5-b]pyridin-6-yl)-2,4-dihydropyrazolo[4,3-b]indole, DIMETHYL SULFOXIDE, ... | | Authors: | Dougan, D.R, Lawson, J.D. | | Deposit date: | 2016-04-19 | | Release date: | 2016-05-25 | | Last modified: | 2016-06-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery of potent, reversible MetAP2 inhibitors via fragment based drug discovery and structure based drug design-Part 2.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
7DIY
 
 | | Crystal structure of SARS-CoV-2 nsp10 bound to nsp14-exoribonuclease domain | | Descriptor: | MAGNESIUM ION, ZINC ION, nsp10 protein, ... | | Authors: | Lin, S, Chen, H, Chen, Z.M, Yang, F.L, Ye, F, Zheng, Y, Yang, J, Lin, X, Sun, H.L, Wang, L.L, Wen, A, Cao, Y, Lu, G.W. | | Deposit date: | 2020-11-19 | | Release date: | 2021-05-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.693 Å) | | Cite: | Crystal structure of SARS-CoV-2 nsp10 bound to nsp14-ExoN domain reveals an exoribonuclease with both structural and functional integrity.
Nucleic Acids Res., 49, 2021
|
|
6KU8
 
 | | structure of HRV-C 3C protein with rupintrivir | | Descriptor: | 4-{2-(4-FLUORO-BENZYL)-6-METHYL-5-[(5-METHYL-ISOXAZOLE-3-CARBONYL)-AMINO]-4-OXO-HEPTANOYLAMINO}-5-(2-OXO-PYRROLIDIN-3-YL)-PENTANOIC ACID ETHYL ESTER, Genome polyprotein | | Authors: | Zhu, L, Yuan, S. | | Deposit date: | 2019-08-31 | | Release date: | 2020-03-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure of the HRV-C 3C-Rupintrivir Complex Provides New Insights for Inhibitor Design.
Virol Sin, 35, 2020
|
|
6KYF
 
 | | Crystal structure of an anti-CRISPR protein | | Descriptor: | AcrF11, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Niu, Y, Wang, H, Zhang, Y, Feng, Y. | | Deposit date: | 2019-09-18 | | Release date: | 2020-09-23 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.07 Å) | | Cite: | A Type I-F Anti-CRISPR Protein Inhibits the CRISPR-Cas Surveillance Complex by ADP-Ribosylation.
Mol.Cell, 80, 2020
|
|
6KU7
 
 | | structure of HRV-C 3C protein | | Descriptor: | Genome polyprotein | | Authors: | Zhu, L, Yuan, S. | | Deposit date: | 2019-08-31 | | Release date: | 2020-03-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure of the HRV-C 3C-Rupintrivir Complex Provides New Insights for Inhibitor Design.
Virol Sin, 35, 2020
|
|
6AOG
 
 | | Crystal structure of Toxoplasma gondii TS-DHFR complexed with NADPH, dUMP, PDDF, and 5-(4-chlorophenyl)-6-ethylpyrimidine-2,4-diamine (pyrimethamine) | | Descriptor: | 10-PROPARGYL-5,8-DIDEAZAFOLIC ACID, 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, 5-(4-CHLORO-PHENYL)-6-ETHYL-PYRIMIDINE-2,4-DIAMINE, ... | | Authors: | Thomas, S.B, Li, Y, Chen, Z, Lu, H. | | Deposit date: | 2017-08-16 | | Release date: | 2018-08-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Identification of selective, brain penetrant Toxoplasma gondii dihydrofolate reductase inhibitors for the treatment of toxoplasmosis
To Be Published
|
|
