6YV4
 
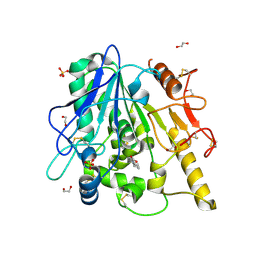 | | STRUCTURE OF THE WNT DEACYLASE NOTUM IN COMPLEX WITH A PYRROLE-3-CARBOXYLIC ACID FRAGMENT 686 | | Descriptor: | 1,2-ETHANEDIOL, 1-cyclopropyl-2,5-dimethyl-pyrrole-3-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Hillier, J, Ruza, R.R, Jones, E.Y. | | Deposit date: | 2020-04-27 | | Release date: | 2020-05-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Screening of a Custom-Designed Acid Fragment Library Identifies 1-Phenylpyrroles and 1-Phenylpyrrolidines as Inhibitors of Notum Carboxylesterase Activity.
J.Med.Chem., 63, 2020
|
|
6YV0
 
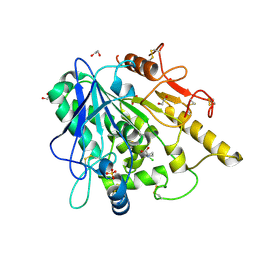 | | STRUCTURE OF THE WNT DEACYLASE NOTUM IN COMPLEX WITH A PYRROLIDINE-3-CARBOXYLIC ACID FRAGMENT 587 | | Descriptor: | (3~{R})-1-(2-chlorophenyl)pyrrolidine-3-carboxylic acid, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ruza, R.R, Hillier, J, Jones, E.Y. | | Deposit date: | 2020-04-27 | | Release date: | 2020-05-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Screening of a Custom-Designed Acid Fragment Library Identifies 1-Phenylpyrroles and 1-Phenylpyrrolidines as Inhibitors of Notum Carboxylesterase Activity.
J.Med.Chem., 63, 2020
|
|
6YUY
 
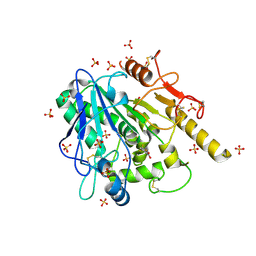 | | STRUCTURE OF THE WNT DEACYLASE NOTUM IN COMPLEX WITH A PYRROLE-3-CARBOXYLIC ACID FRAGMENT 471 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-methyl-5-(trifluoromethyl)-1~{H}-pyrrole-3-carboxylic acid, DIMETHYL SULFOXIDE, ... | | Authors: | Hillier, J, Ruza, R.R, Jones, E.Y. | | Deposit date: | 2020-04-27 | | Release date: | 2020-05-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Screening of a Custom-Designed Acid Fragment Library Identifies 1-Phenylpyrroles and 1-Phenylpyrrolidines as Inhibitors of Notum Carboxylesterase Activity.
J.Med.Chem., 63, 2020
|
|
2WEI
 
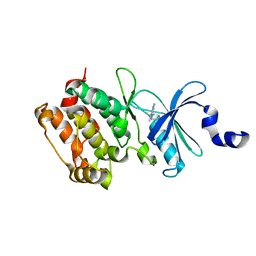 | | Crystal structure of the kinase domain of Cryptosporidium parvum calcium dependent protein kinase in complex with 3-MB-PP1 | | Descriptor: | 1-tert-butyl-3-(3-methylbenzyl)-1H-pyrazolo[3,4-d]pyrimidin-4-amine, CALMODULIN-DOMAIN PROTEIN KINASE 1, PUTATIVE | | Authors: | Roos, A.K, King, O, Chaikuad, A, Zhang, C, Shokat, K.M, Wernimont, A.K, Artz, J.D, Lin, L, MacKenzie, F.I, Finerty, P.J, Vedadi, M, Schapira, M, Indarte, M, Kozieradzki, I, Pike, A.C.W, Fedorov, O, Doyle, D, Muniz, J, Arrowsmith, C.H, Weigelt, J, Edwards, A, Bountra, C, von Delft, F, Heightman, T, Hui, R. | | Deposit date: | 2009-03-31 | | Release date: | 2009-04-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The Cryptosporidium Parvum Kinome.
Bmc Genomics, 12, 2011
|
|
5Y82
 
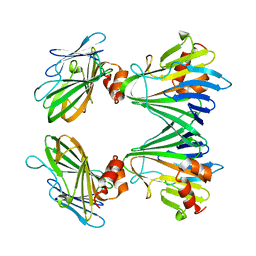 | |
3T0H
 
 | |
3T0Z
 
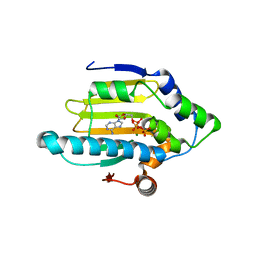 | | Hsp90 N-terminal domain bound to ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Heat shock protein HSP 90-alpha, MAGNESIUM ION | | Authors: | Li, J. | | Deposit date: | 2011-07-21 | | Release date: | 2012-01-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.192 Å) | | Cite: | Structure insights into mechanisms of ATP hydrolysis and the activation of human heat-shock protein 90.
Acta Biochim Biophys Sin (Shanghai), 44, 2012
|
|
6Z43
 
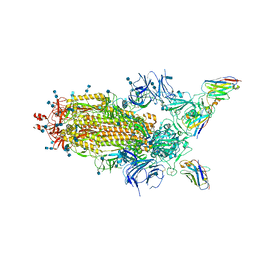 | | Cryo-EM Structure of SARS-CoV-2 Spike : H11-D4 Nanobody Complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Nanobody, ... | | Authors: | Ruza, R.R, Duyvesteyn, H.M.E, Shah, P, Carrique, L, Ren, J, Malinauskas, T, Zhou, D, Stuart, D.I, Naismith, J.H. | | Deposit date: | 2020-05-22 | | Release date: | 2020-06-03 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis for a potent neutralising single-domain antibody that blocks SARS-CoV-2 binding to its receptor ACE2
To Be Published
|
|
6ZH9
 
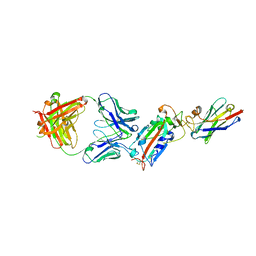 | | Ternary complex CR3022 H11-H4 and RBD (SARS-CoV-2) | | Descriptor: | CR3022 Light chain, CR3022 heavy, Nanobody H11-H4, ... | | Authors: | Naismith, J.H, Mikolajek, H, Le Bas, A. | | Deposit date: | 2020-06-21 | | Release date: | 2020-09-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.31 Å) | | Cite: | Neutralizing nanobodies bind SARS-CoV-2 spike RBD and block interaction with ACE2.
Nat.Struct.Mol.Biol., 27, 2020
|
|
5ZKQ
 
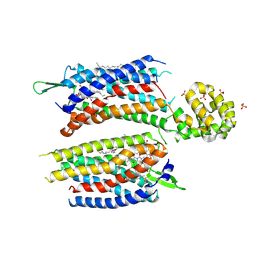 | | Crystal structure of the human platelet-activating factor receptor in complex with ABT-491 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 4-ethynyl-3-{3-fluoro-4-[(2-methyl-1H-imidazo[4,5-c]pyridin-1-yl)methyl]benzene-1-carbonyl}-N,N-dimethyl-1H-indole-1-carboxamide, Platelet-activating factor receptor,Endolysin,Endolysin,Platelet-activating factor receptor, ... | | Authors: | Cao, C, Zhao, Q, Zhang, X.C, Wu, B. | | Deposit date: | 2018-03-25 | | Release date: | 2018-06-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for signal recognition and transduction by platelet-activating-factor receptor.
Nat. Struct. Mol. Biol., 25, 2018
|
|
4NNG
 
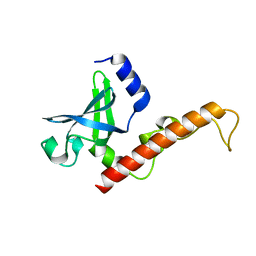 | |
3TYA
 
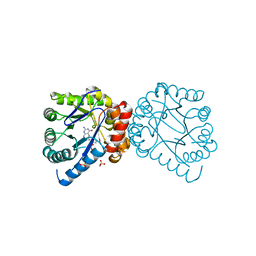 | |
4NNH
 
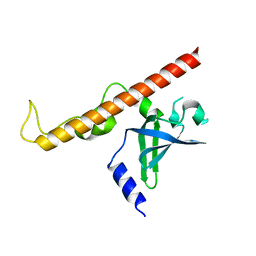 | |
5ZKP
 
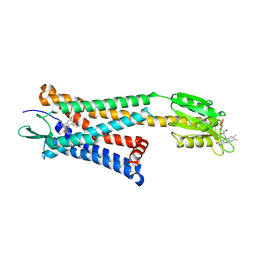 | | Crystal structure of the human platelet-activating factor receptor in complex with SR 27417 | | Descriptor: | FLAVIN MONONUCLEOTIDE, N1,N1-dimethyl-N2-[(pyridin-3-yl)methyl]-N2-{4-[2,4,6-tri(propan-2-yl)phenyl]-1,3-thiazol-2-yl}ethane-1,2-diamine, Platelet-activating factor receptor,Flavodoxin,Platelet-activating factor receptor | | Authors: | Cao, C, Zhao, Q, Zhang, X.C, Wu, B. | | Deposit date: | 2018-03-25 | | Release date: | 2018-06-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Structural basis for signal recognition and transduction by platelet-activating-factor receptor.
Nat. Struct. Mol. Biol., 25, 2018
|
|
3UAS
 
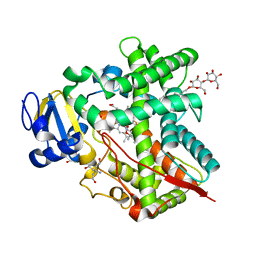 | | Cytochrome P450 2B4 covalently bound to the mechanism-based inactivator 9-ethynylphenanthrene | | Descriptor: | 5-CYCLOHEXYL-1-PENTYL-BETA-D-MALTOSIDE, Cytochrome P450 2B4, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Gay, S.C, Zhang, H, Shah, M.B, Stout, C.D, Halpert, J.R, Hollenberg, P.F. | | Deposit date: | 2011-10-21 | | Release date: | 2013-01-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.939 Å) | | Cite: | Potent Mechanism-Based Inactivation of Cytochrome P450 2B4 by 9-Ethynylphenanthrene: Implications for Allosteric Modulation of Cytochrome P450 Catalysis.
Biochemistry, 52, 2013
|
|
3T10
 
 | | HSP90 N-terminal domain bound to ACP | | Descriptor: | Heat shock protein HSP 90-alpha, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER | | Authors: | Li, J. | | Deposit date: | 2011-07-21 | | Release date: | 2012-01-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Structure insights into mechanisms of ATP hydrolysis and the activation of human heat-shock protein 90.
Acta Biochim Biophys Sin (Shanghai), 44, 2012
|
|
4NNI
 
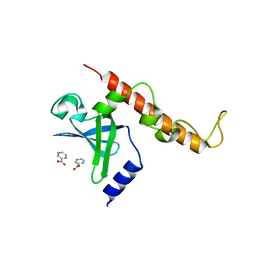 | | Structural basis for targeting the ribosomal protein S1 of Mycobacterium tuberculosis by pyrazinamide | | Descriptor: | 30S ribosomal protein S1, PYRAZINE-2-CARBOXYLIC ACID | | Authors: | Yang, J, Liu, Y, Cai, Q, Lin, D. | | Deposit date: | 2013-11-18 | | Release date: | 2014-12-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Structural basis for targeting the ribosomal protein S1 of Mycobacterium tuberculosis by pyrazinamide.
Mol.Microbiol., 95, 2015
|
|
3TYB
 
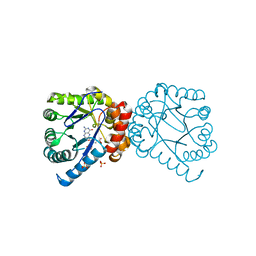 | | Dihydropteroate Synthase in complex with pHBA and DHP+ | | Descriptor: | 2-amino-6-methylidene-6,7-dihydropteridin-4(3H)-one, Dihydropteroate synthase, P-HYDROXYBENZOIC ACID, ... | | Authors: | Yun, M.-K, White, S.W. | | Deposit date: | 2011-09-24 | | Release date: | 2012-03-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Catalysis and sulfa drug resistance in dihydropteroate synthase.
Science, 335, 2012
|
|
4NNK
 
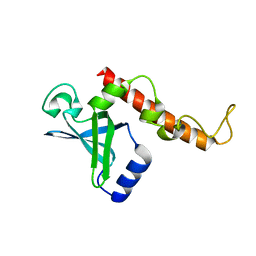 | |
3TYE
 
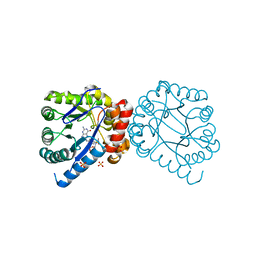 | | Dihydropteroate Synthase in complex with DHP-STZ | | Descriptor: | 2-amino-6-methylidene-6,7-dihydropteridin-4(3H)-one, 4-amino-N-(1,3-thiazol-2-yl)benzenesulfonamide, 4-{[(2-amino-4-oxo-3,4,7,8-tetrahydropteridin-6-yl)methyl]amino}-N-(1,3-thiazol-2-yl)benzenesulfonamide, ... | | Authors: | Yun, M.-K, White, S.W. | | Deposit date: | 2011-09-24 | | Release date: | 2012-03-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Catalysis and sulfa drug resistance in dihydropteroate synthase.
Science, 335, 2012
|
|
3TYU
 
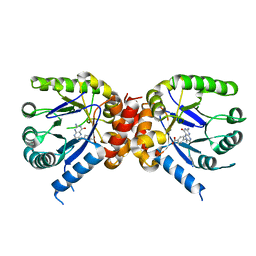 | |
3TYC
 
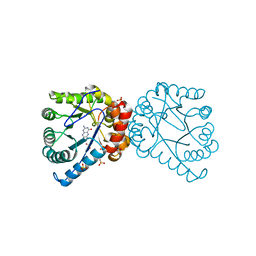 | | Dihydropteroate Synthase in complex with DHP+ | | Descriptor: | 2-amino-6-methylidene-6,7-dihydropteridin-4(3H)-one, Dihydropteroate synthase, SULFATE ION | | Authors: | Yun, M.-K, White, S.W. | | Deposit date: | 2011-09-24 | | Release date: | 2012-03-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.303 Å) | | Cite: | Catalysis and sulfa drug resistance in dihydropteroate synthase.
Science, 335, 2012
|
|
3TYD
 
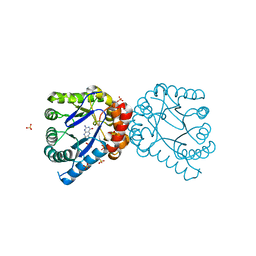 | | Dihydropteroate Synthase in complex with PPi and DHP+ | | Descriptor: | 2-amino-6-methylidene-6,7-dihydropteridin-4(3H)-one, Dihydropteroate synthase, PYROPHOSPHATE 2-, ... | | Authors: | Yun, M.-K, White, S.W. | | Deposit date: | 2011-09-24 | | Release date: | 2012-03-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Catalysis and sulfa drug resistance in dihydropteroate synthase.
Science, 335, 2012
|
|
6AJH
 
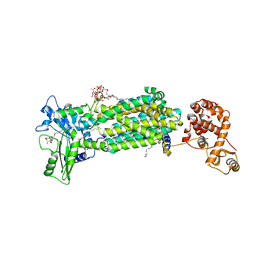 | | Crystal structure of mycolic acid transporter MmpL3 from Mycobacterium smegmatis complexed with AU1235 | | Descriptor: | (CARBAMOYLMETHYL-CARBOXYMETHYL-AMINO)-ACETIC ACID, 1-(2-adamantyl)-3-[2,3,4-tris(fluoranyl)phenyl]urea, Drug exporters of the RND superfamily-like protein,Endolysin, ... | | Authors: | Zhang, B, Li, J, Yang, X.L, Wu, L.J, Yang, H.T, Rao, Z.H. | | Deposit date: | 2018-08-27 | | Release date: | 2018-12-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.818 Å) | | Cite: | Crystal Structures of Membrane Transporter MmpL3, an Anti-TB Drug Target.
Cell, 176, 2019
|
|
6AJI
 
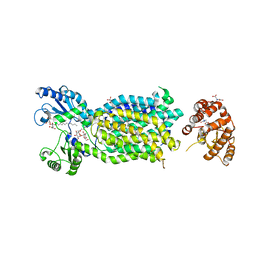 | | Crystal structure of mycolic acid transporter MmpL3 from Mycobacterium smegmatis complexed with Rimonabant | | Descriptor: | (CARBAMOYLMETHYL-CARBOXYMETHYL-AMINO)-ACETIC ACID, 5-(4-chlorophenyl)-1-(2,4-dichlorophenyl)-4-methyl-N-(piperidin-1-yl)-1H-pyrazole-3-carboxamide, Drug exporters of the RND superfamily-like protein,Endolysin, ... | | Authors: | Zhang, B, Li, J, Yang, X.L, Wu, L.J, Yang, H.T, Rao, Z.H. | | Deposit date: | 2018-08-27 | | Release date: | 2018-12-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structures of Membrane Transporter MmpL3, an Anti-TB Drug Target.
Cell, 176, 2019
|
|
