7VEG
 
 | | Understanding NH-pi interaction between Gln and Phe | | Descriptor: | peptide, ACETYLAMINO-ACETIC ACID | | Authors: | Fan, S, Xu, F. | | Deposit date: | 2021-09-08 | | Release date: | 2022-10-19 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Structural Achievability of an NH-pi Interaction between Gln and Phe in a Crystal Structure of a Collagen-like Peptide.
Biomolecules, 12, 2022
|
|
1Z9M
 
 | | Crystal Structure of Nectin-like molecule-1 protein Domain 1 | | Descriptor: | GAPA225 | | Authors: | Dong, X, Xu, F, Gong, Y, Gao, J, Lin, P, Chen, T, Peng, Y, Qiang, B, Yuan, J, Peng, X, Rao, Z. | | Deposit date: | 2005-04-03 | | Release date: | 2006-02-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of the V Domain of Human Nectin-like Molecule-1/Syncam3/Tsll1/Igsf4b, a Neural Tissue-specific Immunoglobulin-like Cell-Cell Adhesion Molecule
J.Biol.Chem., 281, 2006
|
|
8HJ1
 
 | | GPR21(wt) and Gs complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Chen, B, Lin, X, Xu, F. | | Deposit date: | 2022-11-22 | | Release date: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | Cryo-EM structures of orphan GPR21 signaling complexes.
Nat Commun, 14, 2023
|
|
8HSC
 
 | | Gi bound Orphan GPR20 complex with Fab046 in ligand-free state | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Lin, X, Jiang, S, Xu, F. | | Deposit date: | 2022-12-19 | | Release date: | 2023-03-08 | | Last modified: | 2023-03-15 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | The activation mechanism and antibody binding mode for orphan GPR20.
Cell Discov, 9, 2023
|
|
4GRV
 
 | | The crystal structure of the neurotensin receptor NTS1 in complex with neurotensin (8-13) | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Neurotensin 8-13, Neurotensin receptor type 1, ... | | Authors: | Noinaj, N, White, J.F, Shibata, Y, Love, J, Kloss, B, Xu, F, Gvozdenovic-Jeremic, J, Shah, P, Shiloach, J, Tate, C.G, Grisshammer, R. | | Deposit date: | 2012-08-27 | | Release date: | 2012-10-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.802 Å) | | Cite: | Structure of the agonist-bound neurotensin receptor.
Nature, 490, 2012
|
|
5XGP
 
 | | structure of Sizzled from Xenopus laevis at 2.08 angstroms resolution | | Descriptor: | CHLORIDE ION, SULFATE ION, Secreted Xwnt8 inhibitor sizzled | | Authors: | Liu, H, Li, Z, Xu, F. | | Deposit date: | 2017-04-15 | | Release date: | 2017-08-23 | | Last modified: | 2019-12-04 | | Method: | X-RAY DIFFRACTION (2.077 Å) | | Cite: | The crystal structure of full-length Sizzled from Xenopus laevis yields insights into Wnt-antagonistic function of secreted Frizzled-related proteins
J. Biol. Chem., 292, 2017
|
|
5XNQ
 
 | | Crystal structures of human SALM5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Liu, H, Lin, Z, Xu, F. | | Deposit date: | 2017-05-24 | | Release date: | 2018-01-24 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.802 Å) | | Cite: | Structural basis of SALM5-induced PTP delta dimerization for synaptic differentiation
Nat Commun, 9, 2018
|
|
5Y4M
 
 | | Discoidin domain of human CASPR2 | | Descriptor: | 1,2-ETHANEDIOL, human CASPR2 Disc domain | | Authors: | Liu, H, Xu, F, Zhang, J, Liang, W. | | Deposit date: | 2017-08-04 | | Release date: | 2018-08-08 | | Last modified: | 2019-02-20 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Structural mapping of hot spots within human CASPR2 discoidin domain for autoantibody recognition.
J. Autoimmun., 96, 2019
|
|
5YJH
 
 | | Structural insights into periostin functions | | Descriptor: | CALCIUM ION, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Liu, H, Liu, J, Xu, F. | | Deposit date: | 2017-10-10 | | Release date: | 2018-05-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.957 Å) | | Cite: | Structural characterizations of human periostin dimerization and cysteinylation.
FEBS Lett., 592, 2018
|
|
5YBZ
 
 | |
5YJG
 
 | | Structural insights into periostin functions | | Descriptor: | CALCIUM ION, CHLORIDE ION, CYSTEINE, ... | | Authors: | Liu, H, Liu, J, Xu, F. | | Deposit date: | 2017-10-10 | | Release date: | 2018-05-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.399 Å) | | Cite: | Structural characterizations of human periostin dimerization and cysteinylation.
FEBS Lett., 592, 2018
|
|
5YBY
 
 | | Structure of human Gliomedin | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Liu, H, Lin, Z, Xu, F. | | Deposit date: | 2017-09-05 | | Release date: | 2018-10-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.429 Å) | | Cite: | High resolution structure of human gliomedin
To Be Published
|
|
2FR7
 
 | | Crystal Structure of Cytochrome P450 CYP199A2 | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, putative cytochrome P450 | | Authors: | Rao, Z, Wong, L.L, Xu, F, Bell, S.G. | | Deposit date: | 2006-01-19 | | Release date: | 2007-01-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Crystal structure of CYP199A2, a para-substituted benzoic acid oxidizing cytochrome P450 from Rhodopseudomonas palustris
J.Mol.Biol., 383, 2008
|
|
5XNP
 
 | | Crystal structures of human SALM5 in complex with human PTPdelta | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Liu, H, Lin, Z, Xu, F. | | Deposit date: | 2017-05-24 | | Release date: | 2018-01-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.729 Å) | | Cite: | Structural basis of SALM5-induced PTP delta dimerization for synaptic differentiation
Nat Commun, 9, 2018
|
|
8Y6B
 
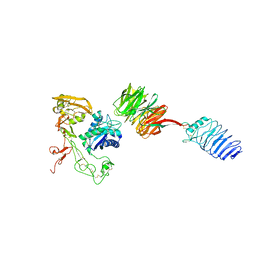 | | Structure of human LGI1-ADAM22 complex in space group P212121 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Disintegrin and metalloproteinase domain-containing protein 22, ... | | Authors: | Liu, H, Xu, F. | | Deposit date: | 2024-02-02 | | Release date: | 2024-02-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.49 Å) | | Cite: | Structure of human LGI1-ADAM22 complex in space group P212121
To Be Published
|
|
5HKE
 
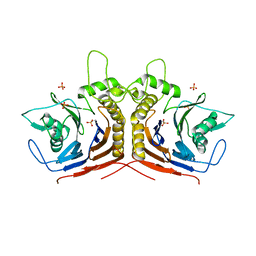 | | bile salt hydrolase from Lactobacillus salivarius | | Descriptor: | Bile salt hydrolase, PHOSPHATE ION | | Authors: | Hu, X.-J. | | Deposit date: | 2016-01-14 | | Release date: | 2016-05-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of bile salt hydrolase from Lactobacillus salivarius.
Acta Crystallogr F Struct Biol Commun, 72, 2016
|
|
5Y7P
 
 | |
6KCW
 
 | | Structure of alginate lyase Aly36B | | Descriptor: | Alginate lyase, CALCIUM ION, PHOSPHATE ION | | Authors: | Dong, F, Chen, X.L, Zhang, Y.Z. | | Deposit date: | 2019-06-29 | | Release date: | 2020-06-24 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Alginate Lyase Aly36B is a New Bacterial Member of the Polysaccharide Lyase Family 36 and Catalyzes by a Novel Mechanism With Lysine as Both the Catalytic Base and Catalytic Acid.
J.Mol.Biol., 431, 2019
|
|
6KHJ
 
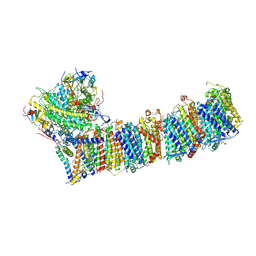 | | Supercomplex for electron transfer | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Pan, X, Cao, D, Xie, F, Zhang, X, Li, M. | | Deposit date: | 2019-07-15 | | Release date: | 2020-02-12 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for electron transport mechanism of complex I-like photosynthetic NAD(P)H dehydrogenase.
Nat Commun, 11, 2020
|
|
1O5T
 
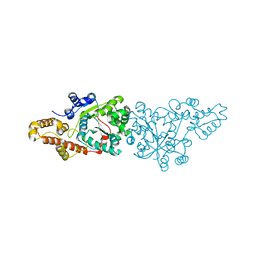 | | Crystal structure of the aminoacylation catalytic fragment of human tryptophanyl-tRNA synthetase | | Descriptor: | Tryptophanyl-tRNA synthetase | | Authors: | Yu, Y, Liu, Y, Shen, N, Xu, X, Jia, J, Jin, Y, Arnold, E, Ding, J. | | Deposit date: | 2003-10-05 | | Release date: | 2004-07-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of Human Tryptophanyl-tRNA Synthetase Catalytic Fragment
J.BIOL.CHEM., 279, 2004
|
|
8W8Q
 
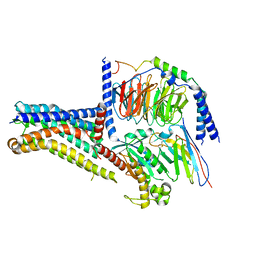 | | Cryo-EM structure of the GPR101-Gs complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Sun, J.P, Gao, N, Yu, X, Wang, G.P, Yang, F, Wang, J.Y, Yang, Z, Guan, Y. | | Deposit date: | 2023-09-04 | | Release date: | 2024-01-03 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Structure of GPR101-Gs enables identification of ligands with rejuvenating potential.
Nat.Chem.Biol., 20, 2024
|
|
8W8S
 
 | | Cryo-EM structure of the AA14-bound GPR101 complex | | Descriptor: | 1-(4-methylpyridin-2-yl)-3-[3-(trifluoromethyl)phenyl]thiourea, Probable G-protein coupled receptor 101 | | Authors: | Sun, J.P, Yu, X, Gao, N, Yang, F, Wang, J.Y, Yang, Z, Guan, Y, Wang, G.P. | | Deposit date: | 2023-09-04 | | Release date: | 2024-01-03 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of GPR101-Gs enables identification of ligands with rejuvenating potential.
Nat.Chem.Biol., 20, 2024
|
|
8W8R
 
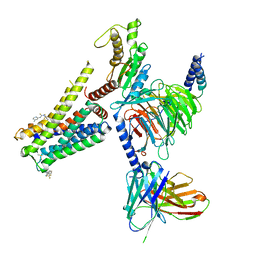 | | Cryo-EM structure of the AA-14-bound GPR101-Gs complex | | Descriptor: | 1-(4-methylpyridin-2-yl)-3-[3-(trifluoromethyl)phenyl]thiourea, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Sun, J.P, Yu, X, Gao, N, Yang, F, Wang, J.Y, Yang, Z, Guan, Y, Wang, G.P. | | Deposit date: | 2023-09-04 | | Release date: | 2024-01-03 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of GPR101-Gs enables identification of ligands with rejuvenating potential.
Nat.Chem.Biol., 20, 2024
|
|
8HJ0
 
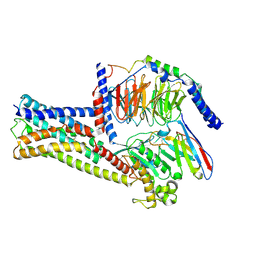 | | GPR21(m5) and G15 complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Chen, B, Lin, X. | | Deposit date: | 2022-11-22 | | Release date: | 2023-03-15 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Cryo-EM structures of orphan GPR21 signaling complexes.
Nat Commun, 14, 2023
|
|
8H0F
 
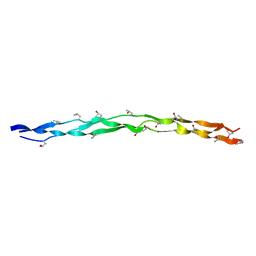 | | Crystal structure of collagen heterotrimer with KD,ER and KE axial pairs | | Descriptor: | collagen-like peptide chain A, collagen-like peptide chain B, collagen-like peptide chain C | | Authors: | Fan, S. | | Deposit date: | 2022-09-28 | | Release date: | 2023-07-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Stability of collagen heterotrimer with same charge pattern and different charged residue identities.
Biophys.J., 122, 2023
|
|
