4W9P
 
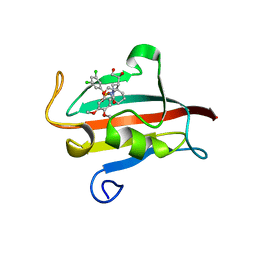 | | The Fk1 domain of FKBP51 in complex with (1S,5S,6R)-10-[(3,5-dichlorophenyl)sulfonyl]-5-[(1S)-1,2-dihydroxyethyl]-3-[2-(3,4-dimethoxyphenoxy)ethyl]-3,10-diazabicyclo[4.3.1]decan-2-one | | Descriptor: | (1S,5S,6R)-10-[(3,5-dichlorophenyl)sulfonyl]-5-[(1S)-1,2-dihydroxyethyl]-3-[2-(3,4-dimethoxyphenoxy)ethyl]-3,10-diazabicyclo[4.3.1]decan-2-one, ACETATE ION, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Pomplun, S, Wang, Y, Kirschner, K, Kozany, C, Bracher, A, Hausch, F. | | Deposit date: | 2014-08-27 | | Release date: | 2014-12-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Rational Design and Asymmetric Synthesis of Potent and Neurotrophic Ligands for FK506-Binding Proteins (FKBPs).
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
2QDW
 
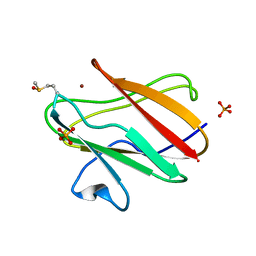 | | Structure of Cu(I) form of the M51A mutant of amicyanin | | Descriptor: | Amicyanin, COPPER (I) ION, PHOSPHATE ION | | Authors: | Ma, J.K, Wang, Y, Carrell, C.J, Mathews, F.S, Davidson, V.L. | | Deposit date: | 2007-06-21 | | Release date: | 2007-12-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (0.92 Å) | | Cite: | A single methionine residue dictates the kinetic mechanism of interprotein electron transfer from methylamine dehydrogenase to amicyanin.
Biochemistry, 46, 2007
|
|
2QTU
 
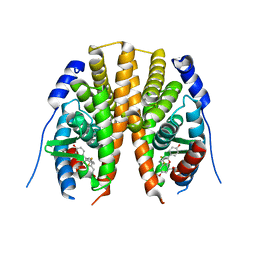 | | Estrogen receptor beta ligand-binding domain complexed to a benzopyran ligand | | Descriptor: | (3aS,4R,9bR)-2,2-difluoro-4-(4-hydroxyphenyl)-6-(methoxymethyl)-1,2,3,3a,4,9b-hexahydrocyclopenta[c]chromen-8-ol, Estrogen receptor beta | | Authors: | Richardson, T.I, Dodge, J.A, Wang, Y, Durbin, J.D, Krishnan, V, Norman, B.H. | | Deposit date: | 2007-08-02 | | Release date: | 2007-10-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Benzopyrans as selective estrogen receptor beta agonists (SERBAs). Part 5: Combined A- and C-ring structure-activity relationship studies.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
6JE9
 
 | | Crystal structure of Nme1Cas9-sgRNA dimer mediated by double protein inhibitor AcrIIC3 monomers | | Descriptor: | AcrIIC3, CRISPR-associated endonuclease Cas9, sgRNA | | Authors: | Sun, W, Yang, J, Cheng, Z, Liu, C, Wang, K, Huang, X, Wang, Y. | | Deposit date: | 2019-02-04 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.46 Å) | | Cite: | Structures of Neisseria meningitidis Cas9 Complexes in Catalytically Poised and Anti-CRISPR-Inhibited States.
Mol.Cell, 76, 2019
|
|
1NA6
 
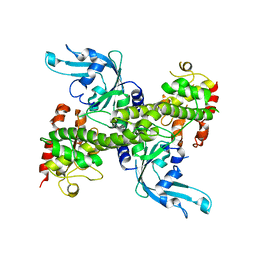 | | Crystal structure of restriction endonuclease EcoRII mutant R88A | | Descriptor: | Restriction endonuclease EcoRII | | Authors: | Zhou, X.E, Wang, Y, Reuter, M, Mucke, M, Kruger, D.H, Meehan, E.J, Chen, L. | | Deposit date: | 2002-11-26 | | Release date: | 2003-12-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of type IIE restriction endonuclease EcoRII reveals an autoinhibition mechanism by a novel effector-binding fold.
J.Mol.Biol., 335, 2004
|
|
6JE4
 
 | | Crystal structure of Nme1Cas9-sgRNA-dsDNA dimer mediated by double protein inhibitor AcrIIC3 monomers | | Descriptor: | 1,2-ETHANEDIOL, AcrIIC3, CRISPR-associated endonuclease Cas9, ... | | Authors: | Sun, W, Yang, J, Cheng, Z, Liu, C, Wang, K, Huang, X, Wang, Y. | | Deposit date: | 2019-02-04 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.069 Å) | | Cite: | Structures of Neisseria meningitidis Cas9 Complexes in Catalytically Poised and Anti-CRISPR-Inhibited States.
Mol.Cell, 76, 2019
|
|
1PCG
 
 | | Helix-stabilized cyclic peptides as selective inhibitors of steroid receptor-coactivator interactions | | Descriptor: | ESTRADIOL, estrogen receptor, peptide inhibitor | | Authors: | Leduc, A.M, Trent, J.O, Wittliff, J.L, Bramlett, K.S, Briggs, S.L, Chirgadze, N.Y, Wang, Y, Burris, T.P, Spatola, A.F. | | Deposit date: | 2003-05-16 | | Release date: | 2003-10-28 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Helix-stabilized cyclic peptides as selective inhibitors of steroid receptor-coactivator interactions
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
5FBJ
 
 | | Complex structure of JMJD5 and substrate | | Descriptor: | (2S)-2-amino-5-[(N-methylcarbamimidoyl)amino]pentanoic acid, 2-OXOGLUTARIC ACID, Lysine-specific demethylase 8, ... | | Authors: | Liu, H.L, Wang, Y, Wang, C, Zhang, G.Y. | | Deposit date: | 2015-12-14 | | Release date: | 2016-12-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | to be published
To Be Published
|
|
6JDQ
 
 | | Crystal structure of Nme1Cas9 in complex with sgRNA | | Descriptor: | CRISPR-associated endonuclease Cas9, sgRNA | | Authors: | Sun, W, Yang, J, Cheng, Z, Liu, C, Wang, K, Huang, X, Wang, Y. | | Deposit date: | 2019-02-02 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structures of Neisseria meningitidis Cas9 Complexes in Catalytically Poised and Anti-CRISPR-Inhibited States.
Mol.Cell, 76, 2019
|
|
6JE3
 
 | | Crystal structure of Nme2Cas9 in complex with sgRNA and target DNA (AGGCCC PAM) with 5 nt overhang | | Descriptor: | 1,2-ETHANEDIOL, CRISPR-associated endonuclease Cas9, non-target DNA strand, ... | | Authors: | Sun, W, Yang, J, Cheng, Z, Liu, C, Wang, K, Huang, X, Wang, Y. | | Deposit date: | 2019-02-03 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.931 Å) | | Cite: | Structures of Neisseria meningitidis Cas9 Complexes in Catalytically Poised and Anti-CRISPR-Inhibited States.
Mol.Cell, 76, 2019
|
|
6JDV
 
 | | Crystal structure of Nme1Cas9 in complex with sgRNA and target DNA (ATATGATT PAM) in catalytic state | | Descriptor: | 1,2-ETHANEDIOL, CRISPR-associated endonuclease Cas9, MAGNESIUM ION, ... | | Authors: | Sun, W, Yang, J, Cheng, Z, Liu, C, Wang, K, Huang, X, Wang, Y. | | Deposit date: | 2019-02-02 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structures of Neisseria meningitidis Cas9 Complexes in Catalytically Poised and Anti-CRISPR-Inhibited States.
Mol.Cell, 76, 2019
|
|
1YT3
 
 | | Crystal Structure of Escherichia coli RNase D, an exoribonuclease involved in structured RNA processing | | Descriptor: | Ribonuclease D, SULFATE ION, ZINC ION | | Authors: | Zuo, Y, Wang, Y, Malhotra, A. | | Deposit date: | 2005-02-09 | | Release date: | 2005-08-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of Escherichia coli RNase D, an Exoribonuclease Involved in Structured RNA Processing
Structure, 13, 2005
|
|
6JFU
 
 | | Crystal structure of Nme2Cas9 in complex with sgRNA and target DNA (AGGCCC PAM) | | Descriptor: | 1,2-ETHANEDIOL, CRISPR-associated endonuclease Cas9, non-target strand, ... | | Authors: | Sun, W, Yang, J, Cheng, Z, Liu, C, Wang, K, Huang, X, Wang, Y. | | Deposit date: | 2019-02-12 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structures of Neisseria meningitidis Cas9 Complexes in Catalytically Poised and Anti-CRISPR-Inhibited States.
Mol.Cell, 76, 2019
|
|
3DZY
 
 | | Intact PPAR gamma - RXR alpha Nuclear Receptor Complex on DNA bound with Rosiglitazone, 9-cis Retinoic Acid and NCOA2 Peptide | | Descriptor: | (9cis)-retinoic acid, 2,4-THIAZOLIDIINEDIONE, 5-[[4-[2-(METHYL-2-PYRIDINYLAMINO)ETHOXY]PHENYL]METHYL]-(9CL), ... | | Authors: | Chandra, V, Huang, P, Hamuro, Y, Raghuram, S, Wang, Y, Burris, T.P, Rastinejad, F. | | Deposit date: | 2008-07-30 | | Release date: | 2008-10-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of the intact PPAR-gamma-RXR- nuclear receptor complex on DNA.
Nature, 456, 2008
|
|
3DZU
 
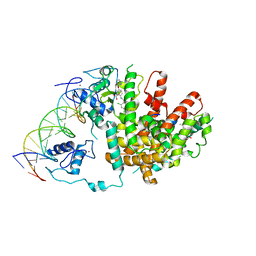 | | Intact PPAR gamma - RXR alpha Nuclear Receptor Complex on DNA bound with BVT.13, 9-cis Retinoic Acid and NCOA2 Peptide | | Descriptor: | (9cis)-retinoic acid, 2-[(2,4-DICHLOROBENZOYL)AMINO]-5-(PYRIMIDIN-2-YLOXY)BENZOIC ACID, DNA (5'-D(*DCP*DAP*DAP*DAP*DCP*DTP*DAP*DGP*DGP*DTP*DCP*DAP*DAP*DAP*DGP*DGP*DTP*DCP*DAP*DG)-3'), ... | | Authors: | Chandra, V, Huang, P, Hamuro, Y, Raghuram, S, Wang, Y, Burris, T.P, Rastinejad, F. | | Deposit date: | 2008-07-30 | | Release date: | 2008-10-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure of the intact PPAR-gamma-RXR- nuclear receptor complex on DNA.
Nature, 456, 2008
|
|
3E00
 
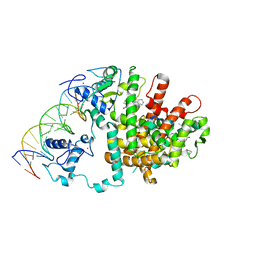 | | Intact PPAR gamma - RXR alpha Nuclear Receptor Complex on DNA bound with GW9662, 9-cis Retinoic Acid and NCOA2 Peptide | | Descriptor: | (9cis)-retinoic acid, 2-chloro-5-nitro-N-phenylbenzamide, DNA (5'-D(*DCP*DAP*DAP*DAP*DCP*DTP*DAP*DGP*DGP*DTP*DCP*DAP*DAP*DAP*DGP*DGP*DTP*DCP*DAP*DG)-3'), ... | | Authors: | Chandra, V, Huang, P, Hamuro, Y, Raghuram, S, Wang, Y, Burris, T.P, Rastinejad, F. | | Deposit date: | 2008-07-30 | | Release date: | 2008-10-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of the intact PPAR-gamma-RXR- nuclear receptor complex on DNA.
Nature, 456, 2008
|
|
1MOZ
 
 | | ADP-ribosylation factor-like 1 (ARL1) from Saccharomyces cerevisiae | | Descriptor: | ADP-ribosylation factor-like protein 1, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Amor, J.C, Horton, J.R, Zhu, X, Wang, Y, Sullards, C, Ringe, D, Cheng, X, Kahn, R.A. | | Deposit date: | 2002-09-10 | | Release date: | 2002-10-09 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (3.17 Å) | | Cite: | Structures of Yeast ARF2 and ARL1:
DISTINCT ROLES FOR THE N TERMINUS IN THE STRUCTURE
AND FUNCTION OF ARF FAMILY GTPases
J.Biol.Chem., 276, 2001
|
|
5V9U
 
 | | Crystal Structure of small molecule ARS-1620 covalently bound to K-Ras G12C | | Descriptor: | (S)-1-{4-[6-chloro-8-fluoro-7-(2-fluoro-6-hydroxyphenyl)quinazolin-4-yl] piperazin-1-yl}propan-1-one, CALCIUM ION, GLYCEROL, ... | | Authors: | Janes, M.R, Zhang, J, Li, L.-S, Hansen, R, Peters, U, Guo, X, Chen, Y, Babbar, A, Firdaus, S.J, Feng, J, Chen, J.H, Li, S, Brehmer, D, Darjania, L, Li, S, Long, Y.O, Thach, C, Liu, Y, Zarieh, A, Ely, T, Kucharski, J.M, Kessler, L.V, Wu, T, Wang, Y, Yao, Y, Deng, X, Zarrinkar, P, Dashyant, D, Lorenzi, M.V, Hu-Lowe, D, Patricelli, M.P, Ren, P, Liu, Y. | | Deposit date: | 2017-03-23 | | Release date: | 2018-02-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Targeting KRAS Mutant Cancers with a Covalent G12C-Specific Inhibitor.
Cell, 172, 2018
|
|
6KC7
 
 | | Crystal structure of Nme1Cas9 in complex with sgRNA and target DNA (ATATGATT PAM) in seed-base paring state | | Descriptor: | CRISPR-associated endonuclease Cas9, DNA (5'-D(*AP*TP*AP*TP*GP*AP*TP*TP*TP*TP*A)-3'), DNA (5'-D(*TP*AP*AP*AP*AP*TP*CP*AP*TP*AP*TP*GP*TP*AP*AP*AP*GP*TP*T)-3'), ... | | Authors: | Sun, W, Yang, J, Cheng, Z, Liu, C, Wang, K, Huang, X, Wang, Y. | | Deposit date: | 2019-06-27 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structures of Neisseria meningitidis Cas9 Complexes in Catalytically Poised and Anti-CRISPR-Inhibited States.
Mol.Cell, 76, 2019
|
|
6KC8
 
 | | Crystal structure of WT Nme1Cas9 in complex with sgRNA and target DNA (ATATGATT PAM) in post-cleavage state | | Descriptor: | CRISPR-associated endonuclease Cas9, DNA (5'-D(*AP*TP*AP*TP*GP*AP*TP*TP*TP*TP*A)-3'), DNA (5'-D(*TP*AP*AP*AP*AP*TP*CP*AP*TP*AP*TP*GP*TP*A)-3'), ... | | Authors: | Sun, W, Yang, J, Cheng, Z, Liu, C, Wang, K, Huang, X, Wang, Y. | | Deposit date: | 2019-06-27 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structures of Neisseria meningitidis Cas9 Complexes in Catalytically Poised and Anti-CRISPR-Inhibited States.
Mol.Cell, 76, 2019
|
|
1PVZ
 
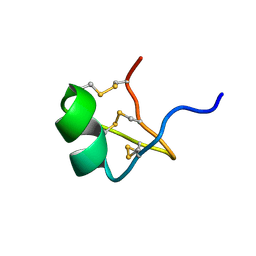 | | Solution Structure of BmP07, A Novel Potassium Channel Blocker from Scorpion Buthus martensi Karsch, 15 structures | | Descriptor: | K+ toxin-like peptide | | Authors: | Wu, H, Zhang, N, Wang, Y, Zhang, Q, Ou, L, Li, M, Hu, G. | | Deposit date: | 2003-06-29 | | Release date: | 2004-05-18 | | Last modified: | 2018-06-20 | | Method: | SOLUTION NMR | | Cite: | Solution structure of BmKK2, a new potassium channel blocker from the venom of chinese scorpion Buthus martensi Karsch
PROTEINS, 55, 2004
|
|
4AUA
 
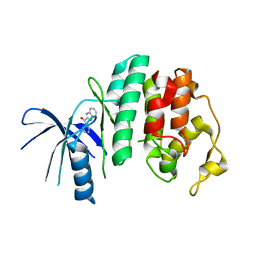 | | Liganded X-ray crystal structure of cyclin dependent kinase 6 (CDK6) | | Descriptor: | 1H-benzimidazol-2-yl(1H-pyrrol-2-yl)methanone, CYCLIN-DEPENDENT KINASE 6 | | Authors: | Cho, Y.S, Angove, H, Brain, C, Chen, C.H.T, Cheng, R, Chopra, R, Chung, K, Congreve, M, Dagostin, C, Davis, D, Feltell, R, Giraldes, J, Hiscock, S, Kim, S, Kovats, S, Lagu, B, Lewry, K, Loo, A, Lu, Y, Luzzio, M, Maniara, W, Mcmenamin, R, Mortenson, P, Benning, R, O'Reilly, M, Rees, D, Shen, J, Smith, T, Wang, Y, Williams, G, Woolford, A, Wrona, W, Xu, M, Yang, F, Howard, S. | | Deposit date: | 2012-05-15 | | Release date: | 2013-02-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Fragment-Based Discovery of 7-Azabenzimidazoles as Potent, Highly Selective, and Orally Active CDK4/6 Inhibitors.
ACS Med Chem Lett, 3, 2012
|
|
1HS7
 
 | | VAM3P N-TERMINAL DOMAIN SOLUTION STRUCTURE | | Descriptor: | SYNTAXIN VAM3 | | Authors: | Dulubova, I, Yamaguchi, T, Wang, Y, Sudhof, T.C, Rizo, J. | | Deposit date: | 2000-12-24 | | Release date: | 2001-03-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Vam3p structure reveals conserved and divergent properties of syntaxins.
Nat.Struct.Biol., 8, 2001
|
|
1I8C
 
 | | SOLUTION STRUCTURE OF THE WATER-SOLUBLE FRAGMENT OF RAT HEPATIC APOCYTOCHROME B5 | | Descriptor: | CYTOCHROME B5 | | Authors: | Falzone, C.J, Wang, Y, Vu, B.C, Scott, N.L, Bhattacharya, S, Lecomte, J.T. | | Deposit date: | 2001-03-13 | | Release date: | 2001-05-16 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural and dynamic perturbations induced by heme binding in cytochrome b5.
Biochemistry, 40, 2001
|
|
2YJY
 
 | | A specific and modular binding code for cytosine recognition in PUF domains | | Descriptor: | 5'-R(*AP*UP*UP*GP*CP*AP*UP*AP*UP*AP)-3', PUMILIO HOMOLOG 1 | | Authors: | Dong, S, Wang, Y, Cassidy-Amstutz, C, Lu, G, Qiu, C, Bigler, R, Jezyk, M, Li, C, Hall, T.M.T, Wang, Z. | | Deposit date: | 2011-05-24 | | Release date: | 2011-06-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.598 Å) | | Cite: | Specific and Modular Binding Code for Cytosine Recognition in Pumilio/Fbf (Puf) RNA-Binding Domains.
J.Biol.Chem., 286, 2011
|
|
