6J23
 
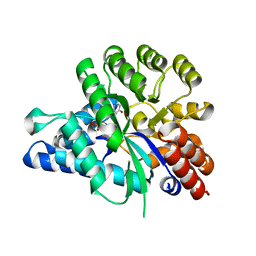 | | Crystal structure of arabidopsis ADAL complexed with GMP | | Descriptor: | Adenosine/AMP deaminase family protein, GUANOSINE-5'-MONOPHOSPHATE, ZINC ION | | Authors: | Wu, B.X, Zhang, D, Nie, H.B, Shen, S.L, Li, S.S, Patel, D.J. | | Deposit date: | 2018-12-30 | | Release date: | 2019-02-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure ofArabidopsis thaliana N6-methyl-AMP deaminase ADAL with bound GMP and IMP and implications forN6-methyl-AMP recognition and processing.
Rna Biol., 16, 2019
|
|
5Y87
 
 | |
5Y85
 
 | |
4ASK
 
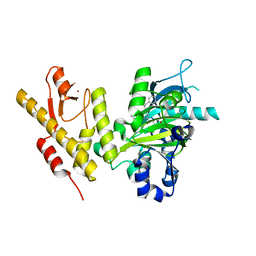 | | CRYSTAL STRUCTURE OF JMJD3 WITH GSK-J1 | | Descriptor: | 3-[[2-pyridin-2-yl-6-(1,2,4,5-tetrahydro-3-benzazepin-3-yl)pyrimidin-4-yl]amino]propanoic acid, COBALT (II) ION, LYSINE-SPECIFIC DEMETHYLASE 6B, ... | | Authors: | Chung, C, Mosley, J, Liddle, J. | | Deposit date: | 2012-05-01 | | Release date: | 2012-07-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | A Selective Jumonji H3K27 Demethylase Inhibitor Modulates the Proinflammatory Macrophage Response
Nature, 488, 2012
|
|
6AF0
 
 | | Structure of Ctr9, Paf1 and Cdc73 ternary complex from Myceliophthora thermophila | | Descriptor: | Cdc73 protein, Ctr9 protein, Paf1 protein | | Authors: | Wang, Z, Deng, P, Zhou, Y. | | Deposit date: | 2018-08-07 | | Release date: | 2018-09-19 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Transcriptional elongation factor Paf1 core complex adopts a spirally wrapped solenoidal topology.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
4N76
 
 | | Structure of Thermus thermophilus Argonaute bound to guide DNA and cleaved target DNA with Mn2+ | | Descriptor: | 5'-D(P*TP*AP*CP*TP*AP*CP*CP*TP*CP*G)-3', 5'-D(P*TP*GP*AP*GP*GP*TP*AP*GP*TP*AP*GP*GP*TP*TP*GP*TP*AP*TP*AP*GP*T)-3', Argonaute, ... | | Authors: | Sheng, G, Zhao, H, Wang, J, Rao, Y, Wang, Y. | | Deposit date: | 2013-10-15 | | Release date: | 2014-01-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Structure-based cleavage mechanism of Thermus thermophilus Argonaute DNA guide strand-mediated DNA target cleavage.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4NCB
 
 | | Structure of Thermus thermophilus Argonaute bound to guide DNA and 19-mer target DNA with Mg2+ | | Descriptor: | 5'-D(*TP*AP*TP*AP*CP*AP*AP*CP*C)-3', 5'-D(*TP*AP*TP*AP*CP*AP*AP*CP*CP*TP*AP*CP*TP*AP*CP*CP*TP*CP*G)-3', 5'-D(P*TP*AP*CP*TP*AP*CP*CP*TP*CP*G)-3', ... | | Authors: | Sheng, G, Zhao, H, Wang, J, Rao, Y, Wang, Y. | | Deposit date: | 2013-10-24 | | Release date: | 2014-01-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.189 Å) | | Cite: | Structure-based cleavage mechanism of Thermus thermophilus Argonaute DNA guide strand-mediated DNA target cleavage.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4KSA
 
 | | Crystal Structure of Malonyl-CoA decarboxylase from Rhodopseudomonas palustris, Northeast Structural Genomics Consortium Target RpR127 | | Descriptor: | MAGNESIUM ION, Malonyl-CoA decarboxylase | | Authors: | Forouhar, F, Neely, H, Seetharaman, J, Sahdev, S, Xiao, R, Patel, D.J, Ciccosanti, C, Wang, D, Everett, J.K, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-05-17 | | Release date: | 2013-06-19 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of malonyl-coenzyme a decarboxylase provide insights into its catalytic mechanism and disease-causing mutations.
Structure, 21, 2013
|
|
4N47
 
 | | Structure of Thermus thermophilus Argonaute bound to guide DNA and 12-mer target DNA | | Descriptor: | 5'-D(*CP*CP*TP*AP*CP*TP*AP*CP*CP*TP*CP*G)-3', 5'-D(P*TP*GP*AP*GP*GP*TP*AP*GP*TP*AP*GP*GP*TP*TP*GP*TP*AP*TP*AP*GP*T)-3', Argonaute, ... | | Authors: | Sheng, G, Zhao, H, Wang, J, Rao, Y, Wang, Y. | | Deposit date: | 2013-10-08 | | Release date: | 2014-01-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.823 Å) | | Cite: | Structure-based cleavage mechanism of Thermus thermophilus Argonaute DNA guide strand-mediated DNA target cleavage.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4NCA
 
 | | Structure of Thermus thermophilus Argonaute bound to guide DNA 19-mer and target DNA in the presence of Mg2+ | | Descriptor: | 5'-D(*AP*CP*AP*AP*CP*C)-3', 5'-D(P*TP*AP*CP*TP*AP*CP*CP*TP*CP*G)-3', 5'-D(P*TP*GP*AP*GP*GP*TP*AP*GP*TP*AP*GP*GP*TP*TP*GP*TP*AP*TP*AP*GP*T)-3', ... | | Authors: | Sheng, G, Zhao, H, Wang, J, Rao, Y, Wang, Y. | | Deposit date: | 2013-10-24 | | Release date: | 2014-01-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.489 Å) | | Cite: | Structure-based cleavage mechanism of Thermus thermophilus Argonaute DNA guide strand-mediated DNA target cleavage.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4N41
 
 | | Structure of Thermus thermophilus Argonaute bound to guide DNA and 15-mer target DNA | | Descriptor: | 5'-D(*AP*AP*CP*CP*TP*AP*CP*TP*GP*CP*CP*TP*CP*G)-3', 5'-D(P*AP*CP*CP*TP*AP*CP*TP*AP*CP*CP*TP*CP*G)-3', 5'-D(P*TP*GP*AP*GP*GP*TP*AP*GP*TP*AP*GP*GP*TP*T*GP*TP*AP*TP*AP*GP*T)-3', ... | | Authors: | Sheng, G, Zhao, H, Wang, J, Rao, Y, Wang, Y. | | Deposit date: | 2013-10-08 | | Release date: | 2014-01-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.248 Å) | | Cite: | Structure-based cleavage mechanism of Thermus thermophilus Argonaute DNA guide strand-mediated DNA target cleavage.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4QBS
 
 | |
4QBR
 
 | |
7LMM
 
 | |
7LMK
 
 | |
4TN7
 
 | | Crystal structure of mouse KDM2A-H3K36ME-NO complex | | Descriptor: | FE (III) ION, Lysine-specific demethylase 2A, NITRIC OXIDE, ... | | Authors: | Cheng, Z. | | Deposit date: | 2014-06-03 | | Release date: | 2015-05-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A molecular threading mechanism underlies Jumonji lysine demethylase KDM2A regulation of methylated H3K36.
Genes Dev., 28, 2014
|
|
2LA5
 
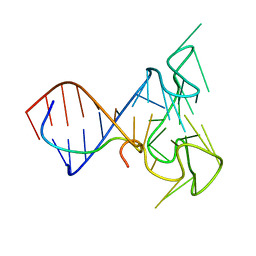 | | RNA Duplex-Quadruplex Junction Complex with FMRP RGG peptide | | Descriptor: | Fragile X mental retardation 1 protein, RNA (36-MER) | | Authors: | Phan, A, Kuryavyi, V, Darnell, J, Serganov, A, Majumdar, A, Ilin, S, Darnell, R, Patel, D. | | Deposit date: | 2011-03-03 | | Release date: | 2011-06-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure-function studies of FMRP RGG peptide recognition of an RNA duplex-quadruplex junction.
Nat.Struct.Mol.Biol., 18, 2011
|
|
3DIM
 
 | |
3DJ2
 
 | |
3DIG
 
 | |
3DIX
 
 | |
3DIR
 
 | |
3DIL
 
 | |
3DJ0
 
 | |
3DIS
 
 | |
