9B4I
 
 | | Filament of D-TLKIVWI, a D-peptide that disaggregates Alzheimer's Paired Helical Filaments, determined by Cryo-EM | | Descriptor: | DTH-DLE-DLY-DIL-DVA-DTR-DIL | | Authors: | Hou, K, Ge, P, Sawaya, M.R, Eisenberg, D.S. | | Deposit date: | 2024-03-20 | | Release date: | 2025-03-12 | | Last modified: | 2025-07-23 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | How short peptides disassemble tau fibrils in Alzheimer's disease.
Nature, 2025
|
|
9B4N
 
 | | Filament of Tau in complex with D-TLKIVWR, a D-peptide that disaggregates Alzheimer's Paired Helical Filaments, determined by Cryo-EM | | Descriptor: | Microtubule-associated protein tau | | Authors: | Hou, K, Ge, P, Sawaya, M.R, Eisenberg, D.S. | | Deposit date: | 2024-03-21 | | Release date: | 2025-03-12 | | Last modified: | 2025-07-23 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | How short peptides disassemble tau fibrils in Alzheimer's disease.
Nature, 2025
|
|
9B4M
 
 | | Filament of Tau in complex with D-TLKIVWS, a D-peptide that disaggregates Alzheimer's Paired Helical Filaments, determined by Cryo-EM | | Descriptor: | Microtubule-associated protein tau | | Authors: | Hou, K, Ge, P, Sawaya, M.R, Eisenberg, D.S. | | Deposit date: | 2024-03-20 | | Release date: | 2025-03-12 | | Last modified: | 2025-07-23 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | How short peptides disassemble tau fibrils in Alzheimer's disease.
Nature, 2025
|
|
9B4L
 
 | | Filament of Tau in complex with D-TLKIVWI, a D-peptide that disaggregates Alzheimer's Paired Helical Filaments, determined by Cryo-EM | | Descriptor: | DTH-DLE-DLY-DIL-DVA-DTR-DIL, Microtubule-associated protein tau, {[-(BIS-CARBOXYMETHYL-AMINO)-ETHYL]-CARBOXYMETHYL-AMINO}-ACETIC ACID | | Authors: | Hou, K, Ge, P, Sawaya, M.R, Eisenberg, D.S. | | Deposit date: | 2024-03-20 | | Release date: | 2025-03-12 | | Last modified: | 2025-07-23 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | How short peptides disassemble tau fibrils in Alzheimer's disease.
Nature, 2025
|
|
7UPE
 
 | | Tau Paired Helical Filament from Alzheimer's Disease not incubated with EGCG | | Descriptor: | Isoform Tau-F of Microtubule-associated protein tau | | Authors: | Seidler, P.M, Murray, K.A, Boyer, D.R, Ge, P, Sawaya, M.R, Eisenberg, D.S. | | Deposit date: | 2022-04-15 | | Release date: | 2022-09-28 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure-based discovery of small molecules that disaggregate Alzheimer's disease tissue derived tau fibrils in vitro.
Nat Commun, 13, 2022
|
|
7UPF
 
 | | Tau Paired Helical Filament from Alzheimer's Disease incubated 1 hr. with EGCG | | Descriptor: | Isoform Tau-F of Microtubule-associated protein tau | | Authors: | Seidler, P.M, Murray, K.A, Boyer, D.R, Ge, P, Sawaya, M.R, Eisenberg, D.S. | | Deposit date: | 2022-04-15 | | Release date: | 2022-09-28 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure-based discovery of small molecules that disaggregate Alzheimer's disease tissue derived tau fibrils in vitro.
Nat Commun, 13, 2022
|
|
7UPG
 
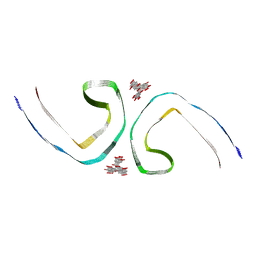 | | Tau Paired Helical Filament from Alzheimer's Disease incubated with EGCG for 3 hours | | Descriptor: | (2R,3R)-5,7-dihydroxy-2-(3,4,5-trihydroxyphenyl)-3,4-dihydro-2H-chromen-3-yl 3,4,5-trihydroxybenzoate, Isoform Tau-F of Microtubule-associated protein tau | | Authors: | Seidler, P.M, Murray, K.A, Boyer, D.R, Ge, P, Sawaya, M.R, Eisenberg, D.S. | | Deposit date: | 2022-04-15 | | Release date: | 2022-09-28 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure-based discovery of small molecules that disaggregate Alzheimer's disease tissue derived tau fibrils in vitro.
Nat Commun, 13, 2022
|
|
5BU3
 
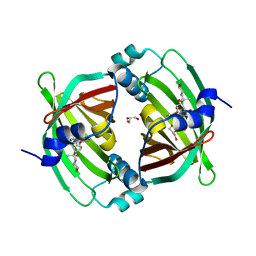 | | Crystal Structure of Diels-Alderase PyrI4 in complex with its product | | Descriptor: | (4S,4aS,6aS,8R,9R,10aR,13R,14aS,18aR,18bR)-9-ethyl-4,8,19-trihydroxy-10a,12,13,18a-tetramethyl-2,3,4,4a,5,6,6a,7,8,9,10,10a,13,14,18a,18b-hexadecahydro-1H-14a,17-(metheno)benzo[b]naphtho[2,1-h]azacyclododecine-16,18(15H,17H)-dione, GLYCEROL, PyrI4 | | Authors: | Pan, L, Guo, Y, Liu, J. | | Deposit date: | 2015-06-03 | | Release date: | 2016-02-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.897 Å) | | Cite: | Enzyme-Dependent [4 + 2] Cycloaddition Depends on Lid-like Interaction of the N-Terminal Sequence with the Catalytic Core in PyrI4
Cell Chem Biol, 23, 2016
|
|
5BTU
 
 | |
4XIC
 
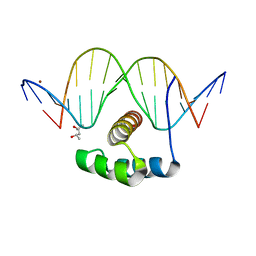 | | ANTPHD WITH 15BP di-thioate modified DNA DUPLEX | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, DNA (5'-D(*AP*GP*AP*AP*AP*GP*CP*(C2S)P*AP*TP*TP*AP*GP*AP*G)-3'), DNA (5'-D(*TP*CP*TP*CP*TP*AP*AP*TP*GP*GP*CP*TP*TP*TP*C)-3'), ... | | Authors: | White, M.A, Zandarashvili, L, Iwahara, J. | | Deposit date: | 2015-01-06 | | Release date: | 2015-11-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Entropic Enhancement of Protein-DNA Affinity by Oxygen-to-Sulfur Substitution in DNA Phosphate.
Biophys.J., 109, 2015
|
|
4X9J
 
 | | EGR-1 with Doubly Methylated DNA | | Descriptor: | DNA (5'-D(*AP*GP*(5CM)P*GP*TP*GP*GP*GP*(5CM)P*GP*T)-3'), DNA (5'-D(*TP*AP*(5CM)P*GP*CP*CP*CP*AP*(5CM)P*GP*C)-3'), Early growth response protein 1, ... | | Authors: | White, M.A, Zandarashvili, L, Iwahara, J. | | Deposit date: | 2014-12-11 | | Release date: | 2015-05-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.412 Å) | | Cite: | Structural impact of complete CpG methylation within target DNA on specific complex formation of the inducible transcription factor Egr-1.
Febs Lett., 589, 2015
|
|
5EOF
 
 | | Crystal structure of OPTN NTD and TBK1 CTD complex | | Descriptor: | Optineurin, Serine/threonine-protein kinase TBK1 | | Authors: | Li, F, Xie, X, Liu, J, Pan, L. | | Deposit date: | 2015-11-10 | | Release date: | 2016-09-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural insights into the interaction and disease mechanism of neurodegenerative disease-associated optineurin and TBK1 proteins.
Nat Commun, 7, 2016
|
|
5EP6
 
 | | The crystal structure of NAP1 in complex with TBK1 | | Descriptor: | 5-azacytidine-induced protein 2, GLYCEROL, Serine/threonine-protein kinase TBK1 | | Authors: | Li, F, Xie, X, Liu, J, Pan, L. | | Deposit date: | 2015-11-11 | | Release date: | 2016-09-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.451 Å) | | Cite: | Structural insights into the interaction and disease mechanism of neurodegenerative disease-associated optineurin and TBK1 proteins.
Nat Commun, 7, 2016
|
|
5VGU
 
 | |
5EOA
 
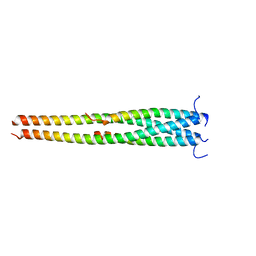 | | Crystal structure of OPTN E50K mutant and TBK1 complex | | Descriptor: | Optineurin, Serine/threonine-protein kinase TBK1 | | Authors: | Li, F, Xie, X, Liu, J, Pan, L. | | Deposit date: | 2015-11-10 | | Release date: | 2016-09-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.503 Å) | | Cite: | Structural insights into the interaction and disease mechanism of neurodegenerative disease-associated optineurin and TBK1 proteins.
Nat Commun, 7, 2016
|
|
6O5V
 
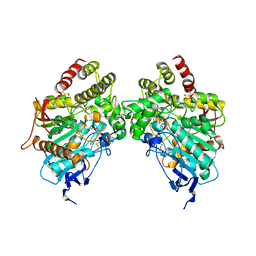 | | Binary complex of native hAChE with oxime reactivator RS-170B | | Descriptor: | 4-carbamoyl-1-(3-{2-[(E)-(hydroxyimino)methyl]-1H-imidazol-1-yl}propyl)pyridin-1-ium, Acetylcholinesterase, GLYCEROL, ... | | Authors: | Gerlits, O, Kovalevsky, A, Radic, Z. | | Deposit date: | 2019-03-04 | | Release date: | 2019-05-29 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.152 Å) | | Cite: | Productive reorientation of a bound oxime reactivator revealed in room temperature X-ray structures of native and VX-inhibited human acetylcholinesterase.
J.Biol.Chem., 294, 2019
|
|
6O66
 
 | |
6O5R
 
 | |
1CKI
 
 | |
1CKJ
 
 | |
6AJH
 
 | | Crystal structure of mycolic acid transporter MmpL3 from Mycobacterium smegmatis complexed with AU1235 | | Descriptor: | (CARBAMOYLMETHYL-CARBOXYMETHYL-AMINO)-ACETIC ACID, 1-(2-adamantyl)-3-[2,3,4-tris(fluoranyl)phenyl]urea, Drug exporters of the RND superfamily-like protein,Endolysin, ... | | Authors: | Zhang, B, Li, J, Yang, X.L, Wu, L.J, Yang, H.T, Rao, Z.H. | | Deposit date: | 2018-08-27 | | Release date: | 2018-12-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.818 Å) | | Cite: | Crystal Structures of Membrane Transporter MmpL3, an Anti-TB Drug Target.
Cell, 176, 2019
|
|
6AJI
 
 | | Crystal structure of mycolic acid transporter MmpL3 from Mycobacterium smegmatis complexed with Rimonabant | | Descriptor: | (CARBAMOYLMETHYL-CARBOXYMETHYL-AMINO)-ACETIC ACID, 5-(4-chlorophenyl)-1-(2,4-dichlorophenyl)-4-methyl-N-(piperidin-1-yl)-1H-pyrazole-3-carboxamide, Drug exporters of the RND superfamily-like protein,Endolysin, ... | | Authors: | Zhang, B, Li, J, Yang, X.L, Wu, L.J, Yang, H.T, Rao, Z.H. | | Deposit date: | 2018-08-27 | | Release date: | 2018-12-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structures of Membrane Transporter MmpL3, an Anti-TB Drug Target.
Cell, 176, 2019
|
|
6AJF
 
 | | Crystal structure of mycolic acid transporter MmpL3 from Mycobacterium smegmatis | | Descriptor: | (CARBAMOYLMETHYL-CARBOXYMETHYL-AMINO)-ACETIC ACID, Drug exporters of the RND superfamily-like protein,Endolysin, alpha-D-glucopyranosyl 6-O-dodecyl-alpha-D-glucopyranoside | | Authors: | Zhang, B, Li, J, Yang, X.L, Wu, L.J, Yang, H.T, Rao, Z.H. | | Deposit date: | 2018-08-27 | | Release date: | 2018-12-26 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.698 Å) | | Cite: | Crystal Structures of Membrane Transporter MmpL3, an Anti-TB Drug Target.
Cell, 176, 2019
|
|
6AJJ
 
 | | Crystal structure of mycolic acid transporter MmpL3 from Mycobacterium smegmatis complexed with ICA38 | | Descriptor: | (CARBAMOYLMETHYL-CARBOXYMETHYL-AMINO)-ACETIC ACID, 4,6-difluoro-N-(spiro[5.5]undecan-3-yl)-1H-indole-2-carboxamide, Drug exporters of the RND superfamily-like protein,Endolysin, ... | | Authors: | Zhang, B, Li, J, Yang, X.L, Wu, L.J, Yang, H.T, Rao, Z.H. | | Deposit date: | 2018-08-27 | | Release date: | 2018-12-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.794 Å) | | Cite: | Crystal Structures of Membrane Transporter MmpL3, an Anti-TB Drug Target.
Cell, 176, 2019
|
|
6AJG
 
 | | Crystal structure of mycolic acid transporter MmpL3 from Mycobacterium smegmatis complexed with SQ109 | | Descriptor: | (CARBAMOYLMETHYL-CARBOXYMETHYL-AMINO)-ACETIC ACID, DODECYL-BETA-D-MALTOSIDE, Drug exporters of the RND superfamily-like protein,Endolysin, ... | | Authors: | Zhang, B, Li, J, Yang, X.L, Wu, L.J, Yang, H.T, Rao, Z.H. | | Deposit date: | 2018-08-27 | | Release date: | 2018-12-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.604 Å) | | Cite: | Crystal Structures of Membrane Transporter MmpL3, an Anti-TB Drug Target.
Cell, 176, 2019
|
|
