4JLH
 
 | |
4GVM
 
 | |
4GW6
 
 | | HIV-1 Integrase Catalytic Core Domain Complexed with Allosteric Inhibitor | | Descriptor: | (2S)-[6-bromo-4-(4-chlorophenyl)-2-methylquinolin-3-yl](tert-butoxy)ethanoic acid, ARSENIC, Gag-Pol polyprotein | | Authors: | Feng, L, Kvaratskhelia, M. | | Deposit date: | 2012-08-31 | | Release date: | 2013-05-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | The A128T Resistance Mutation Reveals Aberrant Protein Multimerization as the Primary Mechanism of Action of Allosteric HIV-1 Integrase Inhibitors.
J.Biol.Chem., 288, 2013
|
|
4WHJ
 
 | |
6MEV
 
 | | Structure of JMJD6 bound to Mono-Methyl Arginine. | | Descriptor: | (2S)-2-amino-5-[(N-methylcarbamimidoyl)amino]pentanoic acid, 2-OXOGLUTARIC ACID, Bifunctional arginine demethylase and lysyl-hydroxylase JMJD6, ... | | Authors: | Lee, S, Zhang, G. | | Deposit date: | 2018-09-07 | | Release date: | 2019-09-18 | | Last modified: | 2020-04-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | JMJD6 cleaves MePCE to release positive transcription elongation factor b (P-TEFb) in higher eukaryotes.
Elife, 9, 2020
|
|
3HPG
 
 | |
3HPH
 
 | | Closed tetramer of Visna virus integrase (residues 1-219) in complex with LEDGF IBD | | Descriptor: | GLYCEROL, Integrase, PC4 and SFRS1-interacting protein, ... | | Authors: | Hare, S, Wang, J, Cherepanov, P. | | Deposit date: | 2009-06-04 | | Release date: | 2009-07-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Structural basis for functional tetramerization of lentiviral integrase
Plos Pathog., 5, 2009
|
|
2ITG
 
 | | CATALYTIC DOMAIN OF HIV-1 INTEGRASE: ORDERED ACTIVE SITE IN THE F185H CONSTRUCT | | Descriptor: | HUMAN IMMUNODEFICIENCY VIRUS-1 INTEGRASE | | Authors: | Bujacz, G, Alexandratos, J, Wlodawer, A, Zhou-Liu, Q, Clement-Mella, C. | | Deposit date: | 1996-09-13 | | Release date: | 1997-03-12 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The catalytic domain of human immunodeficiency virus integrase: ordered active site in the F185H mutant.
FEBS Lett., 398, 1996
|
|
4C0O
 
 | |
4C0Q
 
 | | Transportin 3 in complex with Ran(Q69L)GTP | | Descriptor: | GTP-BINDING NUCLEAR PROTEIN RAN, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Maertens, G, Hare, S, Cherepanov, P. | | Deposit date: | 2013-08-06 | | Release date: | 2014-01-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.42 Å) | | Cite: | Structural Basis for Nuclear Import of Splicing Factors by Human Transportin 3.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4C0P
 
 | | Unliganded Transportin 3 | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, TRANSPORTIN-3 | | Authors: | Maertens, G.N, Cook, N.J, Hare, S, Cherepanov, P. | | Deposit date: | 2013-08-06 | | Release date: | 2014-01-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural Basis for Nuclear Import of Splicing Factors by Human Transportin 3.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4DMN
 
 | | HIV-1 Integrase Catalytical Core Domain | | Descriptor: | (2S)-[6-bromo-4-(4-chlorophenyl)-2-methylquinolin-3-yl](methoxy)ethanoic acid, ARSENIC, HIV-1 Integrase, ... | | Authors: | Feng, L, Kvaratskhelia, M. | | Deposit date: | 2012-02-08 | | Release date: | 2012-03-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Multimode, cooperative mechanism of action of allosteric HIV-1 integrase inhibitors.
J.Biol.Chem., 287, 2012
|
|
1BHL
 
 | | CACODYLATED CATALYTIC DOMAIN OF HIV-1 INTEGRASE | | Descriptor: | HIV-1 INTEGRASE | | Authors: | Maignan, S, Guilloteau, J.P, Zhou-Liu, Q, Clement-Mella, C, Mikol, V. | | Deposit date: | 1998-06-10 | | Release date: | 1998-10-14 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of the catalytic domain of HIV-1 integrase free and complexed with its metal cofactor: high level of similarity of the active site with other viral integrases.
J.Mol.Biol., 282, 1998
|
|
1BI4
 
 | | CATALYTIC DOMAIN OF HIV-1 INTEGRASE | | Descriptor: | INTEGRASE | | Authors: | Maignan, S, Guilloteau, J.P, Zhou-Liu, Q, Clement-Mella, C, Mikol, V. | | Deposit date: | 1998-06-22 | | Release date: | 1998-11-04 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of the catalytic domain of HIV-1 integrase free and complexed with its metal cofactor: high level of similarity of the active site with other viral integrases.
J.Mol.Biol., 282, 1998
|
|
1BL3
 
 | | CATALYTIC DOMAIN OF HIV-1 INTEGRASE | | Descriptor: | INTEGRASE, MAGNESIUM ION | | Authors: | Maignan, S, Guilloteau, J.P, Zhou-Liu, Q, Clement-Mella, C, Mikol, V. | | Deposit date: | 1998-07-23 | | Release date: | 1998-09-30 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of the catalytic domain of HIV-1 integrase free and complexed with its metal cofactor: high level of similarity of the active site with other viral integrases.
J.Mol.Biol., 282, 1998
|
|
3L2Q
 
 | | Crystal structure of the Prototype Foamy Virus (PFV) intasome in apo form | | Descriptor: | 5'-D(*AP*TP*TP*GP*TP*CP*AP*TP*GP*GP*AP*AP*TP*TP*TP*TP*GP*TP*A)-3', 5'-D(*TP*AP*CP*AP*AP*AP*AP*TP*TP*CP*CP*AP*TP*GP*AP*CP*A)-3', GLYCEROL, ... | | Authors: | Hare, S, Gupta, S.S, Cherepanov, P. | | Deposit date: | 2009-12-15 | | Release date: | 2010-02-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Retroviral intasome assembly and inhibition of DNA strand transfer
Nature, 464, 2010
|
|
3L2V
 
 | | Crystal structure of the Prototype Foamy Virus (PFV) intasome in complex with manganese and MK0518 (Raltegravir) | | Descriptor: | 5'-D(*AP*TP*TP*GP*TP*CP*AP*TP*GP*GP*AP*AP*TP*TP*TP*TP*GP*TP*A)-3', 5'-D(*TP*AP*CP*AP*AP*AP*AP*TP*TP*CP*CP*AP*TP*GP*AP*CP*A)-3', AMMONIUM ION, ... | | Authors: | Hare, S, Gupta, S.S, Cherepanov, P. | | Deposit date: | 2009-12-15 | | Release date: | 2010-02-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Retroviral intasome assembly and inhibition of DNA strand transfer
Nature, 464, 2010
|
|
3L2W
 
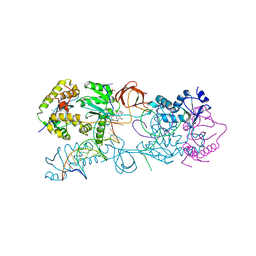 | | Crystal structure of the Prototype Foamy Virus (PFV) intasome in complex with manganese and GS9137 (Elvitegravir) | | Descriptor: | 5'-D(*AP*TP*TP*GP*TP*CP*AP*TP*GP*GP*AP*AP*TP*TP*TP*TP*GP*TP*A)-3', 5'-D(*TP*AP*CP*AP*AP*AP*AP*TP*TP*CP*CP*AP*TP*GP*AP*CP*A)-3', 6-(3-chloro-2-fluorobenzyl)-1-[(1S)-1-(hydroxymethyl)-2-methylpropyl]-7-methoxy-4-oxo-1,4-dihydroquinoline-3-carboxylic acid, ... | | Authors: | Hare, S, Gupta, S.S, Cherepanov, P. | | Deposit date: | 2009-12-15 | | Release date: | 2010-02-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Retroviral intasome assembly and inhibition of DNA strand transfer
Nature, 464, 2010
|
|
3L2U
 
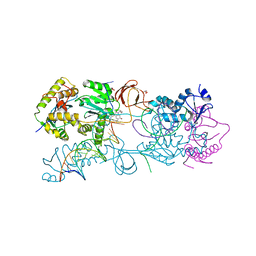 | | Crystal structure of the Prototype Foamy Virus (PFV) intasome in complex with magnesium and GS9137 (Elvitegravir) | | Descriptor: | 5'-D(*AP*TP*TP*GP*TP*CP*AP*TP*GP*GP*AP*AP*TP*TP*TP*TP*GP*TP*A)-3', 5'-D(*TP*AP*CP*AP*AP*AP*AP*TP*TP*CP*CP*AP*TP*GP*AP*CP*A)-3', 6-(3-chloro-2-fluorobenzyl)-1-[(1S)-1-(hydroxymethyl)-2-methylpropyl]-7-methoxy-4-oxo-1,4-dihydroquinoline-3-carboxylic acid, ... | | Authors: | Hare, S, Gupta, S.S, Cherepanov, P. | | Deposit date: | 2009-12-15 | | Release date: | 2010-02-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Retroviral intasome assembly and inhibition of DNA strand transfer
Nature, 464, 2010
|
|
3L2R
 
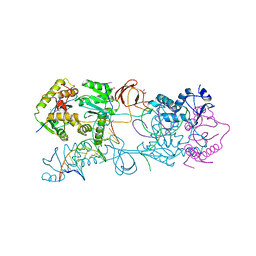 | | Crystal structure of the Prototype Foamy Virus (PFV) intasome in complex with magnesium | | Descriptor: | 5'-D(*AP*TP*TP*GP*TP*CP*AP*TP*GP*GP*AP*AP*TP*TP*TP*TP*GP*TP*A)-3', 5'-D(*TP*AP*CP*AP*AP*AP*AP*TP*TP*CP*CP*AP*TP*GP*AP*CP*A)-3', AMMONIUM ION, ... | | Authors: | Hare, S, Gupta, S.S, Cherepanov, P. | | Deposit date: | 2009-12-15 | | Release date: | 2010-02-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Retroviral intasome assembly and inhibition of DNA strand transfer
Nature, 464, 2010
|
|
3MHP
 
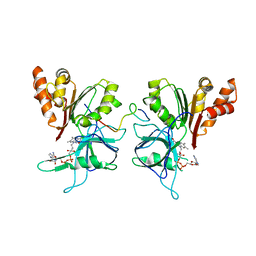 | | FNR-recruitment to the thylakoid | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Ferredoxin--NADP reductase, leaf isozyme, ... | | Authors: | Groll, M, Alte, F, Soll, J, Boelter, B. | | Deposit date: | 2010-04-08 | | Release date: | 2010-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Ferredoxin:NADPH oxidoreductase is recruited to thylakoids by binding to a polyproline type II helix in a pH-dependent manner.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3F9K
 
 | | Two domain fragment of HIV-2 integrase in complex with LEDGF IBD | | Descriptor: | Integrase, MAGNESIUM ION, PC4 and SFRS1-interacting protein, ... | | Authors: | Hare, S, Cherepanov, P. | | Deposit date: | 2008-11-14 | | Release date: | 2009-01-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | A novel co-crystal structure affords the design of gain-of-function lentiviral integrase mutants in the presence of modified PSIP1/LEDGF/p75
Plos Pathog., 5, 2009
|
|
3OY9
 
 | |
1BIS
 
 | | HIV-1 INTEGRASE CORE DOMAIN | | Descriptor: | HIV-1 INTEGRASE | | Authors: | Goldgur, Y, Dyda, F, Hickman, A.B, Jenkins, T.M, Craigie, R, Davies, D.R. | | Deposit date: | 1998-06-19 | | Release date: | 1998-08-19 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Three new structures of the core domain of HIV-1 integrase: an active site that binds magnesium.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
1BIZ
 
 | | HIV-1 INTEGRASE CORE DOMAIN | | Descriptor: | CACODYLATE ION, HIV-1 INTEGRASE | | Authors: | Goldgur, Y, Dyda, F, Hickman, A.B, Jenkins, T.M, Craigie, R, Davies, D.R. | | Deposit date: | 1998-06-21 | | Release date: | 1998-08-19 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Three new structures of the core domain of HIV-1 integrase: an active site that binds magnesium.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
