7FCV
 
 | | Cryo-EM structure of the Potassium channel AKT1 mutant from Arabidopsis thaliana | | Descriptor: | PHOSPHATIDYLETHANOLAMINE, POTASSIUM ION, Potassium channel AKT1 | | Authors: | Yang, G.H, Lu, Y.M, Jia, Y.T, Zhang, Y.M, Tang, R.F, Xu, X, Li, X.M, Lei, J.L. | | Deposit date: | 2021-07-15 | | Release date: | 2022-11-09 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis for the activity regulation of a potassium channel AKT1 from Arabidopsis.
Nat Commun, 13, 2022
|
|
5YS2
 
 | |
5YSL
 
 | | Crystal structure of antibody 1H1 Fab | | Descriptor: | 1H1 heavy chain, 1H1 light chain | | Authors: | Hu, X.L, Yang, F.L. | | Deposit date: | 2017-11-14 | | Release date: | 2017-12-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Two classes of protective antibodies against Pseudorabies virus variant glycoprotein B: Implications for vaccine design.
PLoS Pathog., 13, 2017
|
|
6JHY
 
 | |
6JFW
 
 | | Crystal structure of PA0833 periplasmic domain from Pseudomonas aeruginosa reveals an unexpected enlarged peptidoglycan binding pocket | | Descriptor: | PA0833-PD protein | | Authors: | Lin, X, Ye, F, Lin, S, Yang, F.L, Chen, Z.M, Cao, Y, Chen, Z.J, Gu, J, Lu, G.W. | | Deposit date: | 2019-02-12 | | Release date: | 2019-03-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | Crystal structure of PA0833 periplasmic domain from Pseudomonas aeruginosa reveals an unexpected enlarged peptidoglycan binding pocket.
Biochem. Biophys. Res. Commun., 511, 2019
|
|
8YKY
 
 | | Structure of human class T GPCR TAS2R14-Ggustducin complex with agonist 28.1 | | Descriptor: | 4-methyl-N-[(2M)-2-(1H-tetrazol-5-yl)phenyl]-6-(trifluoromethyl)pyrimidin-2-amine, CHOLESTEROL, G alpha gustducin protein, ... | | Authors: | Hu, X.L, Wu, L.J, Hua, T, Liu, Z.J. | | Deposit date: | 2024-03-05 | | Release date: | 2024-07-10 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Bitter taste TAS2R14 activation by intracellular tastants and cholesterol.
Nature, 631, 2024
|
|
8XQP
 
 | | Structure of human class T GPCR TAS2R14-Gustducin complex with Aristolochic acid A. | | Descriptor: | 8-methoxy-6-nitro-naphtho[1,2-e][1,3]benzodioxole-5-carboxylic acid, CHOLESTEROL, Exo-alpha-sialidase,Taste receptor type 2 member 14,LgBiT, ... | | Authors: | Hu, X.L, Wu, L.J, Hua, T, Liu, Z.J. | | Deposit date: | 2024-01-05 | | Release date: | 2024-07-10 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Bitter taste TAS2R14 activation by intracellular tastants and cholesterol.
Nature, 631, 2024
|
|
8XQN
 
 | | Structure of human class T GPCR TAS2R14-DNGi complex with Aristolochic acid A. | | Descriptor: | 8-methoxy-6-nitro-naphtho[1,2-e][1,3]benzodioxole-5-carboxylic acid, CHOLESTEROL, Exo-alpha-sialidase,Taste receptor type 2 member 14,LgBiT, ... | | Authors: | Hu, X.L, Wu, L.J, Hua, T, Liu, Z.J. | | Deposit date: | 2024-01-05 | | Release date: | 2024-07-10 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Bitter taste TAS2R14 activation by intracellular tastants and cholesterol.
Nature, 631, 2024
|
|
8XQL
 
 | | Structure of human class T GPCR TAS2R14-miniGs/gust complex with Aristolochic acid A. | | Descriptor: | 8-methoxy-6-nitro-naphtho[1,2-e][1,3]benzodioxole-5-carboxylic acid, CHOLESTEROL, Exo-alpha-sialidase,Taste receptor type 2 member 14,LgBiT, ... | | Authors: | Hu, X.L, Wu, L.J, Hua, T, Liu, Z.J. | | Deposit date: | 2024-01-05 | | Release date: | 2024-07-10 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Bitter taste TAS2R14 activation by intracellular tastants and cholesterol.
Nature, 631, 2024
|
|
7C2J
 
 | | Crystal structure of nsp16-nsp10 heterodimer from SARS-CoV-2 in complex with SAM (without additional SAM during crystallization) | | Descriptor: | 2'-O-methyltransferase, Non-structural protein 10, S-ADENOSYLMETHIONINE, ... | | Authors: | Lin, S, Chen, H, Ye, F, Chen, Z.M, Yang, F.L, Zheng, Y, Cao, Y, Qiao, J.X, Yang, S.Y, Lu, G.W. | | Deposit date: | 2020-05-07 | | Release date: | 2020-05-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.799 Å) | | Cite: | Crystal structure of SARS-CoV-2 nsp10/nsp16 2'-O-methylase and its implication on antiviral drug design.
Signal Transduct Target Ther, 5, 2020
|
|
7C2I
 
 | | Crystal structure of nsp16-nsp10 heterodimer from SARS-CoV-2 in complex with SAM (with additional SAM during crystallization) | | Descriptor: | 2'-O-methyltransferase, Non-structural protein 10, S-ADENOSYLMETHIONINE, ... | | Authors: | Lin, S, Chen, H, Ye, F, Chen, Z.M, Yang, F.L, Zheng, Y, Cao, Y, Qiao, J.X, Yang, S.Y, Lu, G.W. | | Deposit date: | 2020-05-07 | | Release date: | 2020-05-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of SARS-CoV-2 nsp10/nsp16 2'-O-methylase and its implication on antiviral drug design.
Signal Transduct Target Ther, 5, 2020
|
|
7DIY
 
 | | Crystal structure of SARS-CoV-2 nsp10 bound to nsp14-exoribonuclease domain | | Descriptor: | MAGNESIUM ION, ZINC ION, nsp10 protein, ... | | Authors: | Lin, S, Chen, H, Chen, Z.M, Yang, F.L, Ye, F, Zheng, Y, Yang, J, Lin, X, Sun, H.L, Wang, L.L, Wen, A, Cao, Y, Lu, G.W. | | Deposit date: | 2020-11-19 | | Release date: | 2021-05-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.693 Å) | | Cite: | Crystal structure of SARS-CoV-2 nsp10 bound to nsp14-ExoN domain reveals an exoribonuclease with both structural and functional integrity.
Nucleic Acids Res., 49, 2021
|
|
7EFP
 
 | | Structure of SARS-CoV-2 spike receptor-binding domain in complex with high affinity ACE2 mutant (S19W,N330Y) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, Spike glycoprotein | | Authors: | Lu, G.W, Ye, F, Lin, X. | | Deposit date: | 2021-03-22 | | Release date: | 2021-11-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.698 Å) | | Cite: | S19W, T27W, and N330Y mutations in ACE2 enhance SARS-CoV-2 S-RBD binding toward both wild-type and antibody-resistant viruses and its molecular basis.
Signal Transduct Target Ther, 6, 2021
|
|
7EFR
 
 | | Structure of SARS-CoV-2 spike receptor-binding domain in complex with high affinity ACE2 mutant (T27W,N330Y) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, Spike glycoprotein | | Authors: | Lu, G.W, Ye, F, Lin, X. | | Deposit date: | 2021-03-23 | | Release date: | 2021-11-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.494 Å) | | Cite: | S19W, T27W, and N330Y mutations in ACE2 enhance SARS-CoV-2 S-RBD binding toward both wild-type and antibody-resistant viruses and its molecular basis.
Signal Transduct Target Ther, 6, 2021
|
|
1J4V
 
 | | CYANOVIRIN-N | | Descriptor: | CYANOVIRIN-N | | Authors: | Clore, G.M, Bewley, C.A. | | Deposit date: | 2001-11-21 | | Release date: | 2002-03-06 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Using conjoined rigid body/torsion angle simulated annealing to determine the relative orientation of covalently linked protein domains from dipolar couplings.
J.Magn.Reson., 154, 2002
|
|
7CKG
 
 | | Crystal structure of TMSiPheRS complexed with TMSiPhe | | Descriptor: | 4-(trimethylsilyl)-L-phenylalanine, Tyrosine--tRNA ligase | | Authors: | Sun, J.P, Wang, J.Y, Zhu, Z.L, He, Q.T, Xiao, P. | | Deposit date: | 2020-07-17 | | Release date: | 2021-03-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.053 Å) | | Cite: | DeSiphering receptor core-induced and ligand-dependent conformational changes in arrestin via genetic encoded trimethylsilyl 1 H-NMR probe.
Nat Commun, 11, 2020
|
|
7CKH
 
 | | Crystal structure of TMSiPheRS | | Descriptor: | Tyrosine--tRNA ligase | | Authors: | Sun, J.P, Wang, J.Y, Zhu, Z.L, He, Q.T, Xiao, P. | | Deposit date: | 2020-07-17 | | Release date: | 2021-03-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.79492676 Å) | | Cite: | DeSiphering receptor core-induced and ligand-dependent conformational changes in arrestin via genetic encoded trimethylsilyl 1 H-NMR probe.
Nat Commun, 11, 2020
|
|
7X4B
 
 | | Crystal Structure of An Anti-CRISPR Protein | | Descriptor: | Anti-CRISPR protein (AcrIIC1), SULFATE ION | | Authors: | Hu, J, Zhang, S, Gao, J.Y, Liu, X, Liu, J. | | Deposit date: | 2022-03-02 | | Release date: | 2022-10-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | A redox switch regulates the assembly and anti-CRISPR activity of AcrIIC1.
Nat Commun, 13, 2022
|
|
6JKM
 
 | | Crystal structure of BubR1 kinase domain | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Lin, L, Ye, S, Huang, Y, Liu, X, Zhang, R, Yao, X. | | Deposit date: | 2019-03-01 | | Release date: | 2019-06-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | BubR1 phosphorylates CENP-E as a switch enabling the transition from lateral association to end-on capture of spindle microtubules.
Cell Res., 29, 2019
|
|
7WSW
 
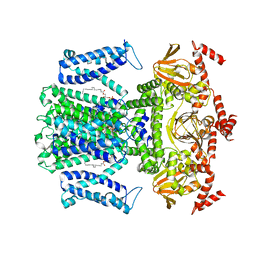 | | Cryo-EM structure of the Potassium channel AKT1 from Arabidopsis thaliana | | Descriptor: | PHOSPHATIDYLETHANOLAMINE, POTASSIUM ION, Potassium channel AKT1 | | Authors: | Yang, G.H, Lu, Y.M, Zhang, Y.M, Jia, Y.T, Li, X.M, Lei, J.L. | | Deposit date: | 2022-02-02 | | Release date: | 2022-11-09 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis for the activity regulation of a potassium channel AKT1 from Arabidopsis.
Nat Commun, 13, 2022
|
|
6JKK
 
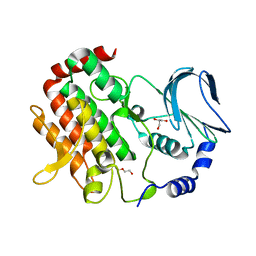 | | Crystal structure of BubR1 kinase domain | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Mitotic checkpoint control protein kinase BUB1 | | Authors: | Lin, L, Ye, S, Huang, Y, Liu, X, Zhang, R, Yao, X. | | Deposit date: | 2019-03-01 | | Release date: | 2019-06-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | BubR1 phosphorylates CENP-E as a switch enabling the transition from lateral association to end-on capture of spindle microtubules.
Cell Res., 29, 2019
|
|
7WRA
 
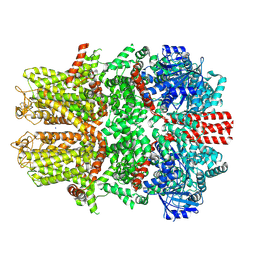 | | Mouse TRPM8 in LMNG in ligand-free state | | Descriptor: | SODIUM ION, Transient receptor potential cation channel subfamily M member 8 | | Authors: | Zhao, C, Xie, Y, Guo, J. | | Deposit date: | 2022-01-26 | | Release date: | 2022-06-22 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Structures of a mammalian TRPM8 in closed state.
Nat Commun, 13, 2022
|
|
7WRF
 
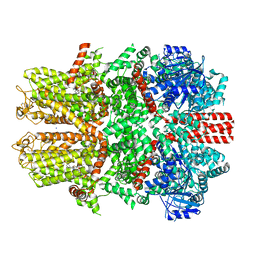 | | Mouse TRPM8 in lipid nanodiscs in the presence of calcium, icilin and PI(4,5)P2 | | Descriptor: | CALCIUM ION, Icilin, SODIUM ION, ... | | Authors: | Zhao, C, Xie, Y, Guo, J. | | Deposit date: | 2022-01-26 | | Release date: | 2022-06-22 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Structures of a mammalian TRPM8 in closed state.
Nat Commun, 13, 2022
|
|
7WRD
 
 | |
7WRC
 
 | | Mouse TRPM8 in LMNG in the presence of calcium, icilin and PI(4,5)P2 | | Descriptor: | CALCIUM ION, Icilin, Transient receptor potential cation channel subfamily M member 8 | | Authors: | Zhao, C, Xie, Y, Guo, J. | | Deposit date: | 2022-01-26 | | Release date: | 2022-06-22 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Structures of a mammalian TRPM8 in closed state.
Nat Commun, 13, 2022
|
|
