6YS3
 
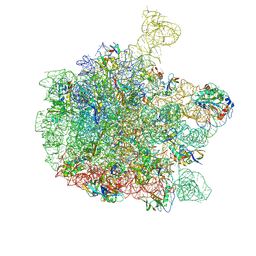 | | Cryo-EM structure of the 50S ribosomal subunit at 2.58 Angstroms with modeled GBC SecM peptide | | Descriptor: | 23S rRNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Schulte, L, Reitz, J, Kudlinzki, D, Hodirnau, V.V, Frangakis, A, Schwalbe, H. | | Deposit date: | 2020-04-20 | | Release date: | 2020-09-30 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.58 Å) | | Cite: | Cryo-EM structure of the 50S ribosomal subunit at 2.58 Angstroms with modeled GBC SecM peptide
Nat Commun, 2020
|
|
8QWJ
 
 | |
8OW2
 
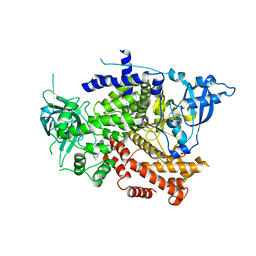 | | Crystal structure of the p110alpha catalytic subunit from homo sapiens in complex with activator 1938 | | Descriptor: | 1-[7-[[2-[[4-(4-ethylpiperazin-1-yl)phenyl]amino]pyridin-4-yl]amino]-2,3-dihydroindol-1-yl]ethanone, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Gong, G.Q, Bellini, D, Vanhaesebroeck, B, Williams, R.L. | | Deposit date: | 2023-04-26 | | Release date: | 2023-05-24 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | A small-molecule PI3K alpha activator for cardioprotection and neuroregeneration.
Nature, 618, 2023
|
|
8PK3
 
 | | CryoEM reconstruction of hemagglutinin HK68 of Influenza A virus bound to an Affimer reagent | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Affimer molecule (A31), ... | | Authors: | Debski-Antoniak, O, Flynn, A, Klebl, D.P, Tiede, C, Muench, S, Tomlinson, D, Fontana, J. | | Deposit date: | 2023-06-24 | | Release date: | 2024-01-03 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Exploiting the Affimer platform against influenza A virus.
Mbio, 15, 2024
|
|
7PKN
 
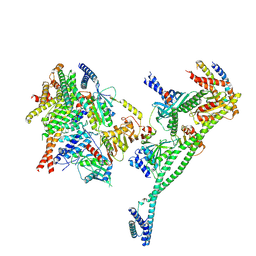 | | Structure of the human CCAN deltaCT complex | | Descriptor: | Centromere protein H, Centromere protein I, Centromere protein K, ... | | Authors: | Muir, K.W, Yatskevich, S, Bellini, D, Barford, D. | | Deposit date: | 2021-08-25 | | Release date: | 2022-04-27 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of the human inner kinetochore bound to a centromeric CENP-A nucleosome.
Science, 376, 2022
|
|
8Q9S
 
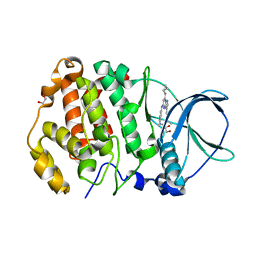 | | STRUCTURE OF PROTEIN KINASE CK2 CATALYTIC SUBUNIT (ISOFORM CK2ALPHA'; CSNK2A2 GENE PRODUCT) IN COMPLEX WITH THE INHIBITOR SGC-CK2-1 | | Descriptor: | 1,2-ETHANEDIOL, Casein kinase II subunit alpha', ~{N}-[5-[[3-cyano-7-(cyclopropylamino)-3~{H}-pyrazolo[1,5-a]pyrimidin-5-yl]amino]-2-methyl-phenyl]propanamide | | Authors: | Werner, C, Lindenblatt, D, Niefind, K. | | Deposit date: | 2023-08-21 | | Release date: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.352 Å) | | Cite: | Discovery and Exploration of Protein Kinase CK2 Binding Sites Using CK2alpha Cys336Ser as an Exquisite Crystallographic Tool
Kinases Phosphatases, 2023
|
|
9FH4
 
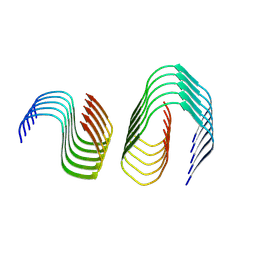 | | Cryo-EM Structure of Amyloid-beta Fibrils Carrying the Uppsala AbetaUpp(1-42)delta(19-24) Mutation - Polymorph 3 | | Descriptor: | Amyloid-beta precursor protein | | Authors: | Zielinski, M, Peralta Reyes, F.S, Gremer, L, Pagnon de la Vega, M, Roeder, C, Heidler, T.V, Syvaenen, S, Willbold, D, Sehlin, D, Ingelsson, M, Schroeder, G.F. | | Deposit date: | 2024-05-26 | | Release date: | 2025-06-11 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Cryo-EM Structures of Amyloid-beta Fibrils from human and murine brains carrying the Uppsala AbetaUpp(1-42)delta(19-24) mutation
To Be Published
|
|
9FH5
 
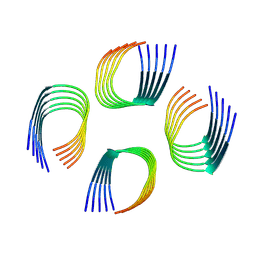 | | Cryo-EM Structure of Amyloid-beta Fibrils Carrying the Uppsala AbetaUpp(1-42)delta(19-24) Mutation - Polymorph 4 | | Descriptor: | Amyloid-beta precursor protein | | Authors: | Zielinski, M, Peralta Reyes, F.S, Gremer, L, Pagnon de la Vega, M, Roeder, C, Heidler, T.V, Syvaenen, S, Willbold, D, Sehlin, D, Ingelsson, M, Schroeder, G.F. | | Deposit date: | 2024-05-26 | | Release date: | 2025-06-11 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM Structures of Amyloid-beta Fibrils from human and murine brains carrying the Uppsala AbetaUpp(1-42)delta(19-24) mutation
To Be Published
|
|
9FH2
 
 | | Cryo-EM Structure of Amyloid-beta Fibrils Carrying the Uppsala AbetaUpp(1-42)delta(19-24) Mutation - Polymorph 1 | | Descriptor: | Amyloid-beta precursor protein | | Authors: | Zielinski, M, Peralta Reyes, F.S, Gremer, L, Pagnon de la Vega, M, Roeder, C, Heidler, T.V, Syvaenen, S, Willbold, D, Sehlin, D, Ingelsson, M, Schroeder, G.F. | | Deposit date: | 2024-05-26 | | Release date: | 2025-06-11 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM Structures of Amyloid-beta Fibrils from human and murine brains carrying the Uppsala AbetaUpp(1-42)delta(19-24) mutation
To Be Published
|
|
9FH1
 
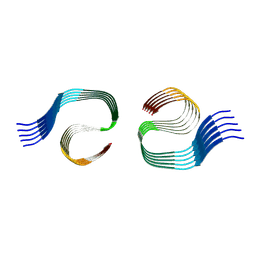 | | Cryo-EM Structure of Amyloid-beta Fibrils from Mouse Brain Carrying the Uppsala AbetaUpp(1-42)delta(19-24) Mutation | | Descriptor: | Amyloid-beta precursor protein | | Authors: | Zielinski, M, Peralta Reyes, F.S, Gremer, L, Pagnon de la Vega, M, Roeder, C, Heidler, T.V, Syvaenen, S, Willbold, D, Sehlin, D, Ingelsson, M, Schroeder, G.F. | | Deposit date: | 2024-05-26 | | Release date: | 2025-06-11 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM Structures of Amyloid-beta Fibrils from human and murine brains carrying the Uppsala AbetaUpp(1-42)delta(19-24) mutation
To Be Published
|
|
9FH6
 
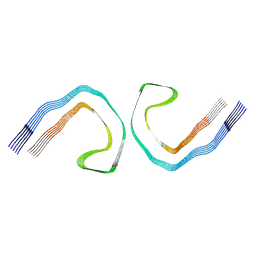 | | Cryo-EM Structure of Tau Filaments from Individuals Carrying the Uppsala AbetaUpp(1-42)delta(19-24) Mutation | | Descriptor: | Microtubule-associated protein tau | | Authors: | Zielinski, M, Peralta Reyes, F.S, Gremer, L, Pagnon de la Vega, M, Roeder, C, Heidler, T.V, Syvaenen, S, Willbold, D, Sehlin, D, Ingelsson, M, Schroeder, G.F. | | Deposit date: | 2024-05-26 | | Release date: | 2025-06-11 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM Structures of Amyloid-beta Fibrils from human and murine brains carrying the Uppsala AbetaUpp(1-42)delta(19-24) mutation
To Be Published
|
|
9FH3
 
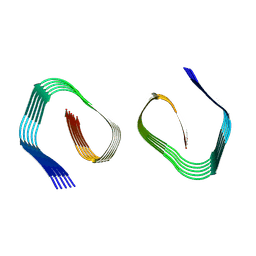 | | Cryo-EM Structure of Amyloid-beta Fibrils Carrying the Uppsala AbetaUpp(1-42)delta(19-24) Mutation - Polymorph 2 | | Descriptor: | Amyloid-beta precursor protein | | Authors: | Zielinski, M, Peralta Reyes, F.S, Gremer, L, Pagnon de la Vega, M, Roeder, C, Heidler, T.V, Syvaenen, S, Willbold, D, Sehlin, D, Ingelsson, M, Schroeder, G.F. | | Deposit date: | 2024-05-26 | | Release date: | 2025-06-11 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM Structures of Amyloid-beta Fibrils from human and murine brains carrying the Uppsala AbetaUpp(1-42)delta(19-24) mutation
To Be Published
|
|
7PII
 
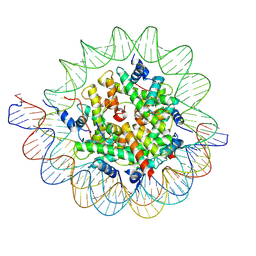 | | Structure of the human CCAN CENP-A alpha-satellite complex | | Descriptor: | Centromere protein C, DNA (122-MER), DNA (123-MER), ... | | Authors: | Yatskevich, S, Muir, K.W, Bellini, D, Barford, D. | | Deposit date: | 2021-08-19 | | Release date: | 2022-05-25 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.68 Å) | | Cite: | Structure of the human inner kinetochore bound to a centromeric CENP-A nucleosome.
Science, 376, 2022
|
|
7R5S
 
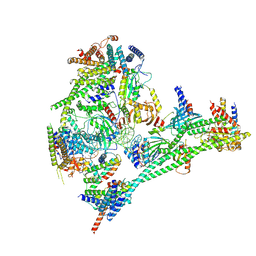 | | Structure of the human CCAN bound to alpha satellite DNA | | Descriptor: | Centromere protein H, Centromere protein I, Centromere protein K, ... | | Authors: | Yatskevich, S, Muir, K.W, Bellini, D, Zhang, Z, Yang, J, Tischer, T, Predin, M, Dendooven, T, McLaughlin, S.H, Barford, D. | | Deposit date: | 2022-02-11 | | Release date: | 2022-04-27 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Structure of the human inner kinetochore bound to a centromeric CENP-A nucleosome.
Science, 376, 2022
|
|
7R5R
 
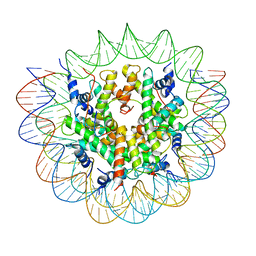 | | Structure of the human CCAN CENP-A alpha-satellite complex | | Descriptor: | Centromere protein C, DNA (171-MER), Histone H2A type 1-C, ... | | Authors: | Yatskevich, S, Muir, K.W, Bellini, D, Zhang, Z, Yang, J, Tischer, T, Predin, M, Dendooven, T, McLaughlin, S.H, Barford, D. | | Deposit date: | 2022-02-11 | | Release date: | 2022-04-27 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.44 Å) | | Cite: | Structure of the human inner kinetochore bound to a centromeric CENP-A nucleosome.
Science, 376, 2022
|
|
7R5V
 
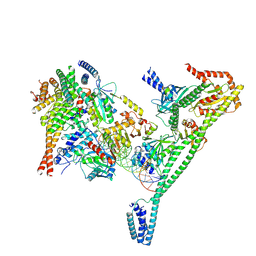 | | Structure of the human CCAN CENP-A alpha-satellite complex | | Descriptor: | Centromere protein H, Centromere protein I, Centromere protein K, ... | | Authors: | Yatskevich, S, Muir, K.W, Bellini, D, Zhang, Z, Yang, J, Tischer, T, Predin, M, Dendooven, T, McLaughlin, S.H, Barford, D. | | Deposit date: | 2022-02-11 | | Release date: | 2022-04-27 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4.55 Å) | | Cite: | Structure of the human inner kinetochore bound to a centromeric CENP-A nucleosome.
Science, 376, 2022
|
|
1XG2
 
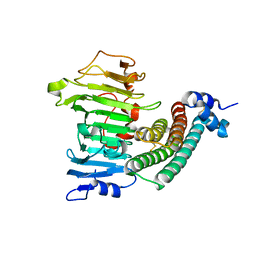 | | Crystal structure of the complex between pectin methylesterase and its inhibitor protein | | Descriptor: | Pectinesterase 1, Pectinesterase inhibitor | | Authors: | Di Matteo, A, Raiola, A, Camardella, L, Giovane, A, Bonivento, D, De Lorenzo, G, Cervone, F, Bellincampi, D, Tsernoglou, D. | | Deposit date: | 2004-09-16 | | Release date: | 2005-03-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis for the Interaction between Pectin Methylesterase and a Specific Inhibitor Protein
Plant Cell, 17, 2005
|
|
1Y8J
 
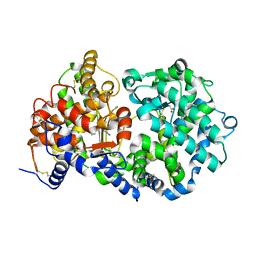 | | Crystal Structure of human NEP complexed with an imidazo[4,5-c]pyridine inhibitor | | Descriptor: | 2-[(1S)-1-BENZYL-2-SULFANYLETHYL]-1H-IMIDAZO[4,5-C]PYRIDIN-5-IUM, 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | Authors: | Sahli, S, Frank, B, Schweizer, W.B, Diederich, F, Blum-Kaelin, D, Aebi, J.D, Bohm, H.J, Oefner, C, Dale, G.E. | | Deposit date: | 2004-12-13 | | Release date: | 2005-06-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Second-Generation Inhibitors for the Metalloprotease Neprilysin Based on Bicyclic Heteroaromatic Scaffolds: Synthesis, Biological Activity, and X-ray Crystal Structure Analysis
HELV.CHIM.ACTA, 88, 2005
|
|
8WVR
 
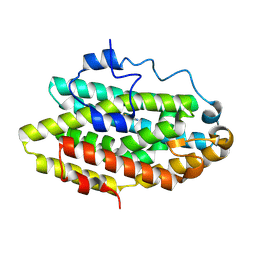 | | Complex structure of the azoxy synthase VlmB and its substrate | | Descriptor: | (2~{S})-2-(2-hexylhydrazinyl)-3-oxidanyl-propanoic acid, FE (III) ION, IMIDAZOLE, ... | | Authors: | Zang, X, Zhou, J.H, Yiling, D, Jingkun, S, Zhijie, Z, Zhuanglin, S, Guiyun, Z. | | Deposit date: | 2023-10-24 | | Release date: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | A conserved enzymatic route for azoxy bond formation in natural product biosynthesis
To Be Published
|
|
7QCK
 
 | | Structure of SARS-CoV-2 Papain-like Protease bound to N-(2,5-dihydroxybenzylidene)-thiosemicarbazone | | Descriptor: | CHLORIDE ION, GLYCEROL, N-(2,5-dihydroxybenzylidene)-thiosemicarbazone, ... | | Authors: | Ewert, W, Gunther, S, Reinke, P, Falke, S, Lieske, J, Miglioli, F, Carcelli, M, Srinivasan, V, Betzel, C, Han, H, Lorenzen, K, Guenther, C, Niebling, S, Garcia-Alai, M, Hinrichs, W, Rogolino, D, Meents, A. | | Deposit date: | 2021-11-24 | | Release date: | 2022-03-16 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Hydrazones and Thiosemicarbazones Targeting Protein-Protein-Interactions of SARS-CoV-2 Papain-like Protease.
Front Chem, 10, 2022
|
|
7QCI
 
 | | Structure of SARS-CoV-2 Papain-like Protease bound to N-(3,4-dihydroxybenzylidene)-thiosemicarbazone | | Descriptor: | CHLORIDE ION, GLYCEROL, N-(3,4-dihydroxybenzylidene)-thiosemicarbazone, ... | | Authors: | Ewert, W, Gunther, S, Reinke, P, Falke, S, Lieske, J, Miglioli, F, Carcelli, M, Srinivasan, V, Betzel, C, Han, H, Lorenzen, K, Guenther, C, Niebling, S, Garcia-Alai, M, Hinrichs, W, Rogolino, D, Meents, A. | | Deposit date: | 2021-11-24 | | Release date: | 2022-03-16 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Hydrazones and Thiosemicarbazones Targeting Protein-Protein-Interactions of SARS-CoV-2 Papain-like Protease.
Front Chem, 10, 2022
|
|
7QCJ
 
 | | Structure of SARS-CoV-2 Papain-like Protease bound to N-(2,4-dihydroxybenzylidene)-thiosemicarbazone | | Descriptor: | CHLORIDE ION, GLYCEROL, N-(2,4-dihydroxybenzylidene)-thiosemicarbazone, ... | | Authors: | Ewert, W, Gunther, S, Reinke, P, Falke, S, Lieske, J, Miglioli, F, Carcelli, M, Srinivasan, V, Betzel, C, Han, H, Lorenzen, K, Guenther, C, Niebling, S, Garcia-Alai, M, Hinrichs, W, Rogolino, D, Meents, A. | | Deposit date: | 2021-11-24 | | Release date: | 2022-03-16 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Hydrazones and Thiosemicarbazones Targeting Protein-Protein-Interactions of SARS-CoV-2 Papain-like Protease.
Front Chem, 10, 2022
|
|
7QCH
 
 | | Structure of SARS-CoV-2 Papain-like Protease bound to N-(3,5-dimethoxy-4-hydroxybenzyliden)thiosemicarbazone | | Descriptor: | CHLORIDE ION, GLYCEROL, N-(3,5-dimetoxy-4-hydroxybenzyliden)thiosemicarbazone, ... | | Authors: | Ewert, W, Gunther, S, Reinke, P, Falke, S, Lieske, J, Miglioli, F, Carcelli, M, Srinivasan, V, Betzel, C, Han, H, Lorenzen, K, Guenther, C, Niebling, S, Garcia-Alai, M, Hinrichs, W, Rogolino, D, Meents, A. | | Deposit date: | 2021-11-24 | | Release date: | 2022-03-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Hydrazones and Thiosemicarbazones Targeting Protein-Protein-Interactions of SARS-CoV-2 Papain-like Protease.
Front Chem, 10, 2022
|
|
7QCM
 
 | | Structure of SARS-CoV-2 Papain-like Protease bound to N-(3-methoxy-4-hydroxy-acetophenone)thiosemicarbazone | | Descriptor: | CHLORIDE ION, GLYCEROL, N-(3-metoxy-4-hydroxy-acetophenone)thiosemicarbazone, ... | | Authors: | Ewert, W, Gunther, S, Reinke, P, Falke, S, Lieske, J, Miglioli, F, Carcelli, M, Srinivasan, V, Betzel, C, Han, H, Lorenzen, K, Guenther, C, Niebling, S, Garcia-Alai, M, Hinrichs, W, Rogolino, D, Meents, A. | | Deposit date: | 2021-11-24 | | Release date: | 2022-03-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Hydrazones and Thiosemicarbazones Targeting Protein-Protein-Interactions of SARS-CoV-2 Papain-like Protease.
Front Chem, 10, 2022
|
|
7QCG
 
 | | Structure of SARS-CoV-2 Papain-like Protease bound to N-(2-pyrrolidyl)-3,4,5-trihydroxybenzoylhydrazone | | Descriptor: | 3,4,5-tris(oxidanyl)-N-[(E)-1H-pyrrol-2-ylmethylideneamino]benzamide, CHLORIDE ION, GLYCEROL, ... | | Authors: | Ewert, W, Gunther, S, Reinke, P, Falke, S, Lieske, J, Miglioli, F, Carcelli, M, Srinivasan, V, Betzel, C, Han, H, Lorenzen, K, Guenther, C, Niebling, S, Garcia-Alai, M, Hinrichs, W, Rogolino, D, Meents, A. | | Deposit date: | 2021-11-23 | | Release date: | 2022-03-23 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Hydrazones and Thiosemicarbazones Targeting Protein-Protein-Interactions of SARS-CoV-2 Papain-like Protease.
Front Chem, 10, 2022
|
|
