8C5I
 
 | | Cyanide dihydratase from Bacillus pumilus C1 variant - Q86R,H305K,H308K,H323K | | Descriptor: | Cyanide dihydratase | | Authors: | Mulelu, A.E, Reitz, J, van Rooyen, J, Scheffer, M, Frangakis, A.S, Dlamini, L.S, Woodward, J.D, Benedik, M.J, Sewell, B.T. | | Deposit date: | 2023-01-09 | | Release date: | 2023-01-18 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | The Role of Histidine Residues in the Oligomerization of Cyanide Dihydratase from Bacillus pumilus C1
To Be Published
|
|
6YS3
 
 | | Cryo-EM structure of the 50S ribosomal subunit at 2.58 Angstroms with modeled GBC SecM peptide | | Descriptor: | 23S rRNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Schulte, L, Reitz, J, Kudlinzki, D, Hodirnau, V.V, Frangakis, A, Schwalbe, H. | | Deposit date: | 2020-04-20 | | Release date: | 2020-09-30 | | Method: | ELECTRON MICROSCOPY (2.58 Å) | | Cite: | Cryo-EM structure of the 50S ribosomal subunit at 2.58 Angstroms with modeled GBC SecM peptide
Nat Commun, 2020
|
|
5M3F
 
 | | Yeast RNA polymerase I elongation complex at 3.8A | | Descriptor: | DNA-directed RNA polymerase I subunit RPA12, DNA-directed RNA polymerase I subunit RPA135, DNA-directed RNA polymerase I subunit RPA14, ... | | Authors: | Neyer, S, Kunz, M, Geiss, C, Hantsche, M, Hodirnau, V.-V, Seybert, A, Engel, C, Scheffer, M.P, Cramer, P, Frangakis, A.S. | | Deposit date: | 2016-10-14 | | Release date: | 2016-11-23 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of RNA polymerase I transcribing ribosomal DNA genes.
Nature, 540, 2016
|
|
5M3M
 
 | | Free monomeric RNA polymerase I at 4.0A resolution | | Descriptor: | DNA-directed RNA polymerase I subunit RPA12, DNA-directed RNA polymerase I subunit RPA135, DNA-directed RNA polymerase I subunit RPA14, ... | | Authors: | Neyer, S, Kunz, M, Geiss, C, Hantsche, M, Hodirnau, V.-V, Seybert, A, Engel, C, Scheffer, M.P, Cramer, P, Frangakis, A.S. | | Deposit date: | 2016-10-15 | | Release date: | 2016-11-23 | | Last modified: | 2019-02-20 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structure of RNA polymerase I transcribing ribosomal DNA genes.
Nature, 540, 2016
|
|
8PBZ
 
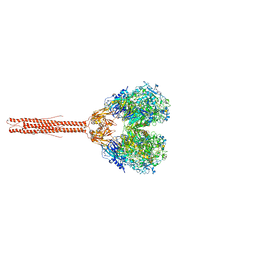 | |
8PC0
 
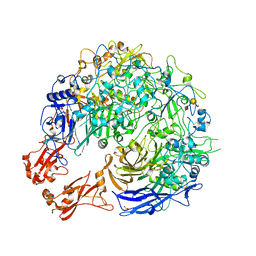 | | Sub-tomogram average of the open conformation of the Nap adhesion complex from the human pathogen Mycoplasma genitalium. | | Descriptor: | Adhesin P1, Mgp-operon protein 3, N-acetyl-alpha-neuraminic acid-(2-6)-beta-D-galactopyranose-(1-4)-1,5-anhydro-D-glucitol, ... | | Authors: | Sprankel, L, Scheffer, M.P, Frangakis, A.F. | | Deposit date: | 2023-06-09 | | Release date: | 2023-11-01 | | Last modified: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (17 Å) | | Cite: | Cryo-electron tomography reveals the binding and release states of the major adhesion complex from Mycoplasma genitalium.
Plos Pathog., 19, 2023
|
|
7A7D
 
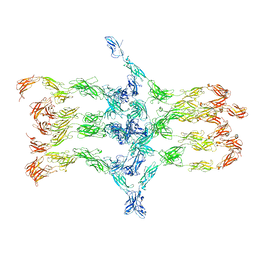 | | Cadherin fit into cryo-ET map | | Descriptor: | Desmocollin-2, Desmoglein-2 | | Authors: | Sikora, M, Ermel, U.H, Seybold, A, Kunz, M, Calloni, G, Reitz, J, Vabulas, R.M, Hummer, G, Frangakis, A.S. | | Deposit date: | 2020-08-28 | | Release date: | 2020-12-23 | | Method: | ELECTRON MICROSCOPY (26 Å) | | Cite: | Desmosome architecture derived from molecular dynamics simulations and cryo-electron tomography.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
8P1I
 
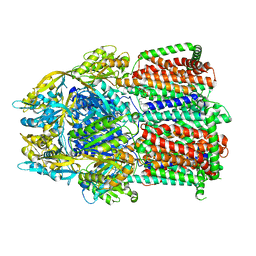 | | Single particle cryo-EM co-structure of Klebsiella pneumoniae AcrB with the BDM91288 efflux pump inhibitor at 2.97 Angstrom resolution | | Descriptor: | 1,2-DIOCTANOYL-SN-GLYCERO-3-[PHOSPHO-RAC-(1-GLYCEROL), 3-chloranyl-2,6-di(piperazin-4-ium-1-yl)quinoline, Efflux pump membrane transporter, ... | | Authors: | Boernsen, C, Mueller, R.T, Pos, K.M, Frangakis, A.S. | | Deposit date: | 2023-05-12 | | Release date: | 2024-01-17 | | Last modified: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Pyridylpiperazine efflux pump inhibitor boosts in vivo antibiotic efficacy against K. pneumoniae.
Embo Mol Med, 16, 2024
|
|
6QDW
 
 | | Cryo-EM structure of the 50S ribosomal subunit at 2.83 Angstroms with modeled GBC SecM peptide | | Descriptor: | 23S rRNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Schulte, L, Reitz, J, Hodirnau, V.V, Kudlinzki, D, Mao, J, Glaubitz, C, Frangakis, A, Schwalbe, H. | | Deposit date: | 2019-01-03 | | Release date: | 2020-01-15 | | Last modified: | 2020-12-02 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Cysteine oxidation and disulfide formation in the ribosomal exit tunnel.
Nat Commun, 11, 2020
|
|
7B8P
 
 | | Acinetobacter baumannii multidrug transporter AdeB in OOO state | | Descriptor: | Efflux pump membrane transporter | | Authors: | Ornik-Cha, A, Reitz, J, Seybert, A, Frangakis, A, Pos, K.M. | | Deposit date: | 2020-12-13 | | Release date: | 2021-10-20 | | Last modified: | 2021-12-08 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | Structural and functional analysis of the promiscuous AcrB and AdeB efflux pumps suggests different drug binding mechanisms.
Nat Commun, 12, 2021
|
|
7B8Q
 
 | | Acinetobacter baumannii multidrug transporter AdeB in L*OO state | | Descriptor: | Efflux pump membrane transporter | | Authors: | Ornik-Cha, A, Reitz, J, Seybert, A, Frangakis, A, Pos, K.M. | | Deposit date: | 2020-12-13 | | Release date: | 2021-10-20 | | Last modified: | 2021-12-08 | | Method: | ELECTRON MICROSCOPY (3.84 Å) | | Cite: | Structural and functional analysis of the promiscuous AcrB and AdeB efflux pumps suggests different drug binding mechanisms.
Nat Commun, 12, 2021
|
|
8PBY
 
 | |
8PC1
 
 | |
8PBX
 
 | |
8P4I
 
 | | Cyanide dihydratase from Bacillus pumilus C1 | | Descriptor: | Cyanide dihydratase | | Authors: | Mulelu, A.E, Reitz, J, van Rooyen, J.M, Scheffer, M, Frangakis, A.S, Dlamini, L.S, Woodward, J.D, Benedik, M.J, Sewell, B.T. | | Deposit date: | 2023-05-22 | | Release date: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (3.83 Å) | | Cite: | The Role of Histidine Residues in the Oligomerization of Cyanide Dihydratase from Bacillus pumilus C1
To Be Published
|
|
