2YBY
 
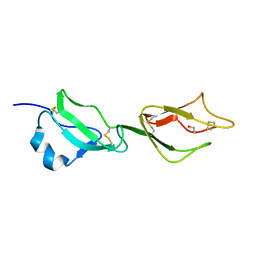 | | Structure of domains 6 and 7 of the mouse complement regulator Factor H | | Descriptor: | 1,2-ETHANEDIOL, COMPLEMENT FACTOR H | | Authors: | Everett, R.J, Caesar, J.J.E, Johnson, S.J, Tang, C.M, Lea, S.M. | | Deposit date: | 2011-03-30 | | Release date: | 2012-04-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Design and Evaluation of Meningococcal Vaccines Through Structure-Based Modification of Host and Pathogen Molecules.
Plos Pathog., 8, 2012
|
|
4QQJ
 
 | |
4QQT
 
 | |
4QRC
 
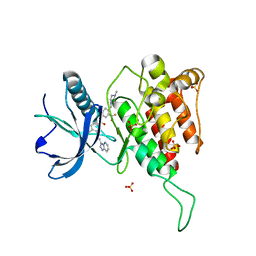 | | Crystal Structure of the Tyrosine Kinase Domain of FGF Receptor 4 in Complex with Ponatinib | | Descriptor: | 3-(imidazo[1,2-b]pyridazin-3-ylethynyl)-4-methyl-N-{4-[(4-methylpiperazin-1-yl)methyl]-3-(trifluoromethyl)phenyl}benzam ide, Fibroblast growth factor receptor 4, SULFATE ION | | Authors: | Huang, Z, Mohammadi, M. | | Deposit date: | 2014-06-30 | | Release date: | 2014-10-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | DFG-out Mode of Inhibition by an Irreversible Type-1 Inhibitor Capable of Overcoming Gate-Keeper Mutations in FGF Receptors.
Acs Chem.Biol., 10, 2015
|
|
5EK5
 
 | | STRUCTURAL CHARACTERIZATION OF IRMA FROM ESCHERICHIA COLI | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Heras, B, Moriel, D.G, Paxman, J.J, Schembri, M.A. | | Deposit date: | 2015-11-03 | | Release date: | 2016-03-09 | | Last modified: | 2016-07-13 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Molecular and Structural Characterization of a Novel Escherichia coli Interleukin Receptor Mimic Protein.
Mbio, 7, 2016
|
|
8F9Y
 
 | | SAL1 from Arabidopsis thaliana | | Descriptor: | MAGNESIUM ION, PHOSPHATE ION, SAL1 phosphatase | | Authors: | Frkic, R.L, Kaczmarski, J.A, Tan, L, Jackson, C.J. | | Deposit date: | 2022-11-24 | | Release date: | 2023-01-18 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Sensing and signaling of oxidative stress in chloroplasts by inactivation of the SAL1 phosphoadenosine phosphatase.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
7E4Z
 
 | | Crystal structure of tubulin in complex with Maytansinol | | Descriptor: | (1R,2S,3S,5S,6S,16E,18E,20R,21S)-11-chloro-6,21-dihydroxy-12,20-dimethoxy-2,5,9,16-tetramethyl-4,24-dioxa-9,22-diazatetracyclo[19.3.1.1~10,14~.0~3,5~]hexacosa-10(26),11,13,16,18-pentaene-8,23-dione, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Wang, Y, Li, W. | | Deposit date: | 2021-02-16 | | Release date: | 2021-07-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | C3 ester side chain plays a pivotal role in the antitumor activity of Maytansinoids.
Biochem.Biophys.Res.Commun., 566, 2021
|
|
7E4Q
 
 | | Crystal structure of tubulin in complex with L-DM1-SMe | | Descriptor: | (1S,2R,3R,5R,6R,16E,18E,20R,21S)-11-chloro-21-hydroxy-12,20-dimethoxy-2,5,9,16-tetramethyl-8,23-dioxo-4,24-dioxa-9,22-diazatetracyclo[19.3.1.1~10,14~.0~3,5~]hexacosa-10(26),11,13,16,18-pentaen-6-yl (2S)-2-{methyl[3-(methyldisulfanyl)propanoyl]amino}propanoate (non-preferred name), 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Wang, Y, Li, W. | | Deposit date: | 2021-02-14 | | Release date: | 2021-07-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | C3 ester side chain plays a pivotal role in the antitumor activity of Maytansinoids.
Biochem.Biophys.Res.Commun., 566, 2021
|
|
7E4R
 
 | | Crystal structure of tubulin in complex with D-DM1-SMe | | Descriptor: | (10E,12E)-86-chloro-14-hydroxy-85,14-dimethoxy-33,2,7,10-tetramethyl-12,6-dioxo-7-aza-1(6,4)-oxazinana-3(2,3)-oxirana-8(1,3)-benzenacyclotetradecaphane-10,12-dien-4-yl N-methyl-N-(3-(methylsulfinothioyl)propanoyl)-D-alaninate, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Wang, Y, Li, W. | | Deposit date: | 2021-02-15 | | Release date: | 2021-07-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.597 Å) | | Cite: | C3 ester side chain plays a pivotal role in the antitumor activity of Maytansinoids.
Biochem.Biophys.Res.Commun., 566, 2021
|
|
7EN3
 
 | | Crystal structure of tubulin in complex with Tubulysin analogue TGL | | Descriptor: | (2S,4R)-5-(4-fluorophenyl)-2-methyl-4-[[2-[(1R,3R)-4-methyl-3-[5-methylhexyl-[(2S,3S)-3-methyl-2-[[(2R)-1-methylpiperidin-2-yl]carbonylamino]pentanoyl]amino]-1-oxidanyl-pentyl]-1,3-thiazol-4-yl]carbonylamino]pentanoic acid, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Wang, Y, Li, W. | | Deposit date: | 2021-04-15 | | Release date: | 2022-04-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.643 Å) | | Cite: | The X-ray structure of tubulysin analogue TGL in complex with tubulin and three possible routes for the development of next-generation tubulysin analogues.
Biochem.Biophys.Res.Commun., 565, 2021
|
|
3ZTX
 
 | | Aurora kinase selective inhibitors identified using a Taxol-induced checkpoint sensitivity screen. | | Descriptor: | 2-((4-(4-HYDROXYPIPERIDIN-1-YL)PHENYL)AMINO)-5,11-DIMETHYL-5H-BENZO[E]PYRIMIDO [5,4-B][1,4]DIAZEPIN-6(11H)-ONE, INNER CENTROMERE PROTEIN A, SERINE/THREONINE-PROTEIN KINASE 12-A | | Authors: | Kwiatkowski, N, Villa, F, Musacchio, A, Gray, N. | | Deposit date: | 2011-07-12 | | Release date: | 2012-02-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Selective Aurora Kinase Inhibitors Identified Using a Taxol- Induced Checkpoint Sensitivity Screen.
Acs Chem.Biol., 7, 2012
|
|
6BEA
 
 | |
6ATE
 
 | | SRC kinase bound to covalent inhibitor | | Descriptor: | N-{2-[(5-chloro-2-{[4-(4-methylpiperazin-1-yl)phenyl]amino}pyrimidin-4-yl)amino]phenyl}propanamide, Proto-oncogene tyrosine-protein kinase Src | | Authors: | Gurbani, D, Westover, K.D. | | Deposit date: | 2017-08-28 | | Release date: | 2019-02-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.402 Å) | | Cite: | Leveraging Compound Promiscuity to Identify Targetable Cysteines within the Kinome.
Cell Chem Biol, 26, 2019
|
|
6DUN
 
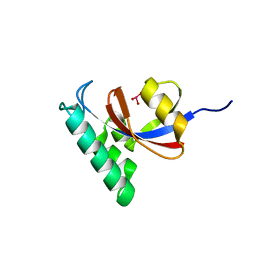 | | Crystal Structure Analysis of PIN1 | | Descriptor: | Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1, TRIHYDROXYARSENITE(III) | | Authors: | Seo, H.-S, Dhe-Paganon, S. | | Deposit date: | 2018-06-21 | | Release date: | 2019-03-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Arsenic targets Pin1 and cooperates with retinoic acid to inhibit cancer-driving pathways and tumor-initiating cells.
Nat Commun, 9, 2018
|
|
4CKR
 
 | | Crystal structure of the human DDR1 kinase domain in complex with DDR1-IN-1 | | Descriptor: | 1,2-ETHANEDIOL, 4-[(4-ethylpiperazin-1-yl)methyl]-n-{4-methyl-3-[(2-oxo-2,3-dihydro-1h-indol-5-yl)oxy]phenyl}-3-(trifluoromethyl)benzamide, EPITHELIAL DISCOIDIN DOMAIN-CONTAINING RECEPTOR 1 | | Authors: | Canning, P, Elkins, J.M, Goubin, S, Mahajan, P, Krojer, T, Newman, J.A, Dixon-Clarke, S, Chaikuad, A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A. | | Deposit date: | 2014-01-07 | | Release date: | 2014-01-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of a Potent and Selective Ddr1 Receptor Tyrosine Kinase Inhibitor.
Acs Chem.Biol., 8, 2013
|
|
6DAU
 
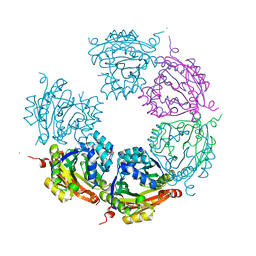 | | Crystal structure of E33Q and E41Q mutant forms of the spermidine/spermine N-acetyltransferase SpeG from Vibrio cholerae | | Descriptor: | GLYCEROL, Spermidine N1-acetyltransferase | | Authors: | Filippova, E.V, Minasov, G, Beahan, A, Kulyavtsev, P, Tan, L, Tran, D, Kuhn, M.L, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-05-02 | | Release date: | 2018-07-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Crystal structure of E33Q and E41Q mutant forms of the spermidine/spermine N-acetyltransferase SpeG from Vibrio cholerae.
To be Published
|
|
5Y2T
 
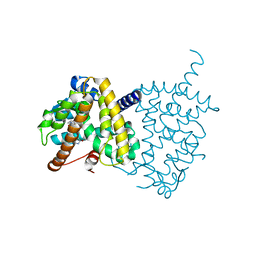 | | Structure of PPARgamma ligand binding domain - lobeglitazone complex | | Descriptor: | (5S)-5-[[4-[2-[[6-(4-methoxyphenoxy)pyrimidin-4-yl]-methyl-amino]ethoxy]phenyl]methyl]-1,3-thiazolidine-2,4-dione, Peroxisome proliferator-activated receptor gamma | | Authors: | Im, Y.J, Lee, M. | | Deposit date: | 2017-07-27 | | Release date: | 2017-12-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structures of PPAR gamma complexed with lobeglitazone and pioglitazone reveal key determinants for the recognition of antidiabetic drugs
Sci Rep, 7, 2017
|
|
5Y2O
 
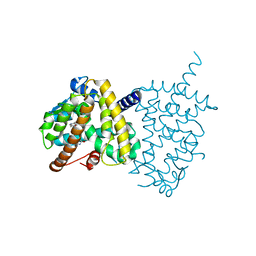 | | Structure of PPARgamma ligand binding domain-pioglitazone complex | | Descriptor: | (5S)-5-[[4-[2-(5-ethylpyridin-2-yl)ethoxy]phenyl]methyl]-1,3-thiazolidine-2,4-dione, Peroxisome proliferator-activated receptor gamma | | Authors: | Im, Y.J, Lee, M. | | Deposit date: | 2017-07-26 | | Release date: | 2017-12-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Structures of PPAR gamma complexed with lobeglitazone and pioglitazone reveal key determinants for the recognition of antidiabetic drugs
Sci Rep, 7, 2017
|
|
6IYB
 
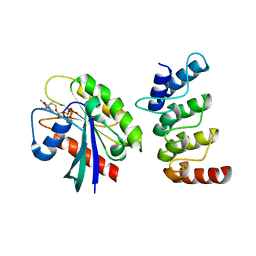 | | Structure of human ORP1 ANK - Rab7 complex | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Oxysterol-binding protein-related protein 1, ... | | Authors: | Tong, J, Im, Y.J. | | Deposit date: | 2018-12-14 | | Release date: | 2019-03-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.091 Å) | | Cite: | Structural basis of human ORP1-Rab7 interaction for the late-endosome and lysosome targeting.
PLoS ONE, 14, 2019
|
|
8HF2
 
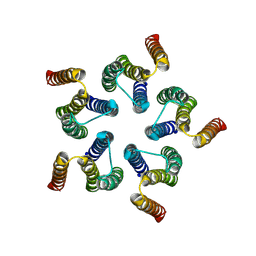 | | Cryo-EM structure of WeiTsing | | Descriptor: | PRA1 family protein | | Authors: | Qin, L, Tang, L.H, Chen, Y.H. | | Deposit date: | 2022-11-09 | | Release date: | 2023-06-21 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (4.14 Å) | | Cite: | WeiTsing, a pericycle-expressed ion channel, safeguards the stele to confer clubroot resistance.
Cell, 186, 2023
|
|
8GUG
 
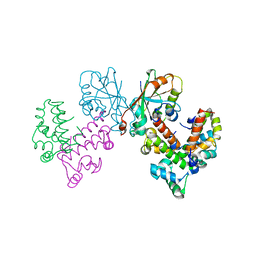 | | Structure of VPA0770 toxin bound to VPA0769 antitoxin in Vibrio parahaemolyticus | | Descriptor: | DUF2384 domain-containing protein, RES domain-containing protein | | Authors: | Song, X.J, Zhang, Y, Xu, Y.Y, Lin, Z. | | Deposit date: | 2022-09-12 | | Release date: | 2023-07-26 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural insights of the toxin-antitoxin system VPA0770-VPA0769 in Vibrio parahaemolyticus.
Int.J.Biol.Macromol., 242, 2023
|
|
8H59
 
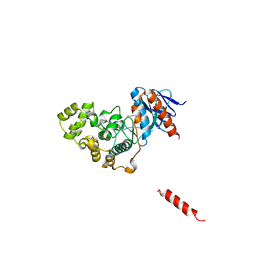 | | A fungal MAP kinase in complex with an inhibitor | | Descriptor: | Mitogen-activated protein kinase MPS1, ~{N}-[(2~{S})-3-(1~{H}-indol-3-yl)-1-(methylamino)-1-oxidanylidene-propan-2-yl]-8-[2-methoxy-5-(trifluoromethyloxy)phenyl]-1,6-naphthyridine-2-carboxamide | | Authors: | Kong, Z, Zhang, X, Wang, D, Liu, J. | | Deposit date: | 2022-10-12 | | Release date: | 2023-02-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure-Aided Identification of an Inhibitor Targets Mps1 for the Management of Plant-Pathogenic Fungi.
Mbio, 14, 2023
|
|
8HFY
 
 | | SARS-CoV-2 Omicron BA.1 spike protein receptor-binding domain in complex with white-tailed deer ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, ... | | Authors: | Han, P, Meng, Y.M, Qi, J.X. | | Deposit date: | 2022-11-13 | | Release date: | 2023-08-30 | | Last modified: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Structural basis of white-tailed deer, Odocoileus virginianus , ACE2 recognizing all the SARS-CoV-2 variants of concern with high affinity.
J.Virol., 97, 2023
|
|
8HFX
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron BA.1 spike protein in complex with white-tailed deer ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, ... | | Authors: | Han, P, Meng, Y.M, Qi, J.X. | | Deposit date: | 2022-11-13 | | Release date: | 2023-08-30 | | Last modified: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Structural basis of white-tailed deer, Odocoileus virginianus , ACE2 recognizing all the SARS-CoV-2 variants of concern with high affinity.
J.Virol., 97, 2023
|
|
8HG0
 
 | | Cryo-EM structure of SARS-CoV-2 prototype spike protein receptor-binding domain in complex with white-tailed deer ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, Spike protein S1, ... | | Authors: | Han, P, Meng, Y.M, Qi, J.X. | | Deposit date: | 2022-11-13 | | Release date: | 2023-08-30 | | Last modified: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (3.51 Å) | | Cite: | Structural basis of white-tailed deer, Odocoileus virginianus , ACE2 recognizing all the SARS-CoV-2 variants of concern with high affinity.
J.Virol., 97, 2023
|
|
