1DOF
 
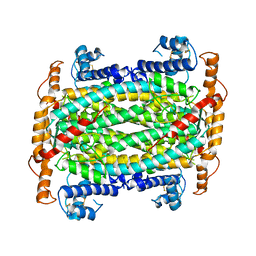 | | THE CRYSTAL STRUCTURE OF ADENYLOSUCCINATE LYASE FROM PYROBACULUM AEROPHILUM: INSIGHTS INTO THERMAL STABILITY AND HUMAN PATHOLOGY | | Descriptor: | ADENYLOSUCCINATE LYASE | | Authors: | Toth, E.A, Yeates, T.O, Goedken, E, Dixon, J.E, Marqusee, S. | | Deposit date: | 1999-12-20 | | Release date: | 2001-01-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of adenylosuccinate lyase from Pyrobaculum aerophilum reveals an intracellular protein with three disulfide bonds.
J.Mol.Biol., 301, 2000
|
|
1F1O
 
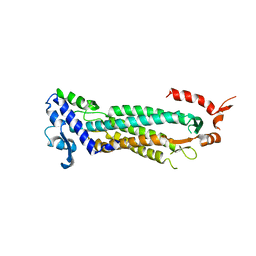 | | STRUCTURAL STUDIES OF ADENYLOSUCCINATE LYASES | | Descriptor: | ADENYLOSUCCINATE LYASE | | Authors: | Toth, E.A, Yeates, T. | | Deposit date: | 2000-05-19 | | Release date: | 2001-01-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | The crystal structure of adenylosuccinate lyase from Pyrobaculum aerophilum reveals an intracellular protein with three disulfide bonds.
J.Mol.Biol., 301, 2000
|
|
1C3C
 
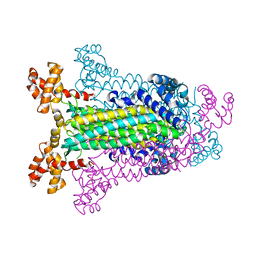 | | T. MARITIMA ADENYLOSUCCINATE LYASE | | Descriptor: | PROTEIN (ADENYLOSUCCINATE LYASE) | | Authors: | Toth, E.A, Yeates, T.O. | | Deposit date: | 1999-07-27 | | Release date: | 2000-02-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structure of adenylosuccinate lyase, an enzyme with dual activity in the de novo purine biosynthetic pathway.
Structure Fold.Des., 8, 2000
|
|
1C3U
 
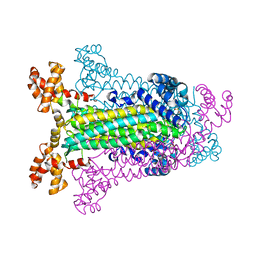 | | T. MARITIMA ADENYLOSUCCINATE LYASE | | Descriptor: | ADENYLOSUCCINATE LYASE, SULFATE ION | | Authors: | Toth, E.A, Yeates, T.O. | | Deposit date: | 1999-07-28 | | Release date: | 2000-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The structure of adenylosuccinate lyase, an enzyme with dual activity in the de novo purine biosynthetic pathway.
Structure Fold.Des., 8, 2000
|
|
1Q57
 
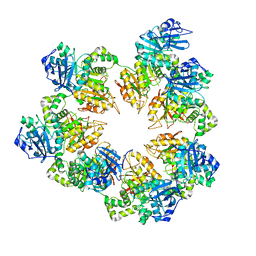 | | The Crystal Structure of the Bifunctional Primase-Helicase of Bacteriophage T7 | | Descriptor: | DNA primase/helicase | | Authors: | Toth, E.A, Li, Y, Sawaya, M.R, Cheng, Y, Ellenberger, T. | | Deposit date: | 2003-08-06 | | Release date: | 2003-11-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | The Crystal Structure of the Bifunctional Primase-Helicase of Bacteriophage T7
Mol.Cell, 12, 2003
|
|
3N5N
 
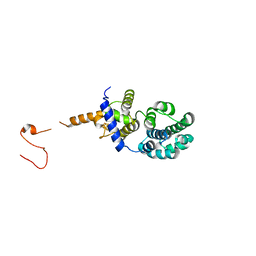 | |
6UAI
 
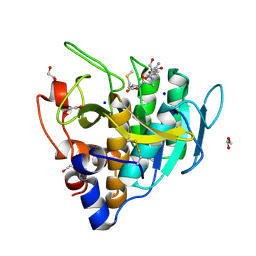 | | Imidazole-triggered RAS-specific subtilisin SUBT_BACAM complexed with YSAM peptide | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Toth, E.A, Bryan, P.N, Orban, J. | | Deposit date: | 2019-09-10 | | Release date: | 2020-09-16 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Engineering subtilisin proteases that specifically degrade active RAS.
Commun Biol, 4, 2021
|
|
6UAO
 
 | | Imidazole-triggered RAS-specific subtilisin SUBT_BACAM complexed with the peptide EEYSAM | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Peptide EEYSAM, ... | | Authors: | Toth, E.A, Bryan, P.N, Orban, J. | | Deposit date: | 2019-09-11 | | Release date: | 2020-09-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Engineering subtilisin proteases that specifically degrade active RAS.
Commun Biol, 4, 2021
|
|
6UBE
 
 | | Azide-triggered subtilisin SUBT_BACAM complexed with the peptide LFRAL | | Descriptor: | AZIDE ION, GLYCEROL, Peptide LFRAL, ... | | Authors: | Toth, E.A, Bryan, P.N, Orban, J, Gallagher, D.T, Custer, G. | | Deposit date: | 2019-09-11 | | Release date: | 2020-09-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Engineering subtilisin proteases that specifically degrade active RAS.
Commun Biol, 4, 2021
|
|
6U9L
 
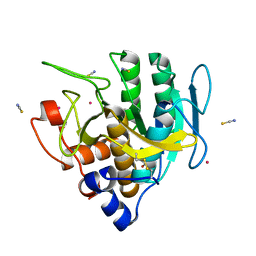 | | Imidazole-triggered RAS-specific subtilisin SUBT_BACAM | | Descriptor: | GLYCEROL, POTASSIUM ION, SUBTILISIN BPN', ... | | Authors: | Toth, E.A, Bryan, P.N, Orban, J. | | Deposit date: | 2019-09-09 | | Release date: | 2020-09-16 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Engineering subtilisin proteases that specifically degrade active RAS.
Commun Biol, 4, 2021
|
|
5TKQ
 
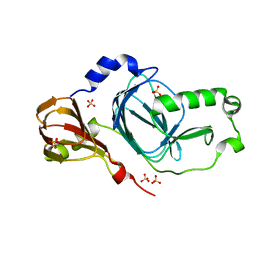 | | Crystal structure of human 3HAO with zinc bound in the active site | | Descriptor: | 3-hydroxyanthranilate 3,4-dioxygenase, SULFATE ION, ZINC ION | | Authors: | Pidugu, L.S, Toth, E.A. | | Deposit date: | 2016-10-07 | | Release date: | 2017-04-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structures of human 3-hydroxyanthranilate 3,4-dioxygenase with native and non-native metals bound in the active site.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
5TK5
 
 | | Crystal structure of human 3HAO with iron bound in the active site | | Descriptor: | 3-hydroxyanthranilate 3,4-dioxygenase, FE (III) ION, SULFATE ION | | Authors: | Pidugu, L.S, Toth, E.A. | | Deposit date: | 2016-10-06 | | Release date: | 2017-04-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Crystal structures of human 3-hydroxyanthranilate 3,4-dioxygenase with native and non-native metals bound in the active site.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
5EA2
 
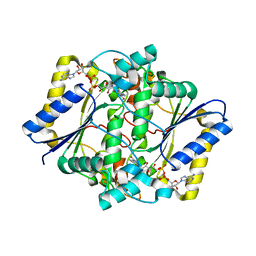 | | Crystal Structure of Holo NAD(P)H dehydrogenase, quinone 1 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NAD(P)H dehydrogenase [quinone] 1 | | Authors: | Pidugu, L.S, Mbimba, J.E, Ahmad, M, Pozharski, E, Sausville, E.A, Emadi, A, Toth, E.A. | | Deposit date: | 2015-10-15 | | Release date: | 2016-02-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | A direct interaction between NQO1 and a chemotherapeutic dimeric naphthoquinone.
Bmc Struct.Biol., 16, 2016
|
|
5EAI
 
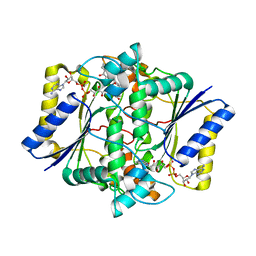 | | Crystal Structure of NAD(P)H dehydrogenase, quinone 1 complexed with a chemotherapeutic naphthoquinone E6a | | Descriptor: | (2~{R},3~{R})-2-[(2~{S},3~{S})-3-bromanyl-1,4-bis(oxidanylidene)-2,3-dihydronaphthalen-2-yl]-3-oxidanyl-2,3-dihydronaphthalene-1,4-dione, FLAVIN-ADENINE DINUCLEOTIDE, NAD(P)H dehydrogenase [quinone] 1 | | Authors: | Pidugu, L.S, Mbimba, J.E, Ahmad, M, Pozharski, E, Sausville, E.A, Emadi, A, Toth, E.A. | | Deposit date: | 2015-10-16 | | Release date: | 2016-02-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A direct interaction between NQO1 and a chemotherapeutic dimeric naphthoquinone.
Bmc Struct.Biol., 16, 2016
|
|
5DKQ
 
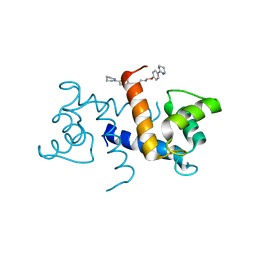 | | Crystal Structure of Calcium-loaded S100B bound to SBi4214 | | Descriptor: | 2,2'-[pentane-1,5-diylbis(oxybenzene-4,1-diyl)]di-1,4,5,6-tetrahydropyrimidine, CALCIUM ION, Protein S100-B | | Authors: | Cavalier, M.C, Ansari, M.I, Pierce, A.D, Wilder, P.T, McKnight, L.E, Raman, E.P, Neau, D.B, Bezawada, P, Alasady, M.J, Varney, K.M, Toth, E.A, MacKerell Jr, A.D, Coop, A, Weber, D.J. | | Deposit date: | 2015-09-03 | | Release date: | 2016-01-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.591 Å) | | Cite: | Small Molecule Inhibitors of Ca(2+)-S100B Reveal Two Protein Conformations.
J.Med.Chem., 59, 2016
|
|
5DKR
 
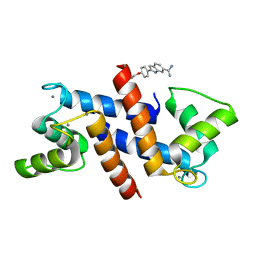 | | Crystal Structure of Calcium-loaded S100B bound to SBi29 | | Descriptor: | 2-[4-(4-carbamimidoylphenoxy)phenyl]-1H-indole-6-carboximidamide, CALCIUM ION, Protein S100-B | | Authors: | Cavalier, M.C, Ansari, M.I, Pierce, A.D, Wilder, P.T, McKnight, L.E, Raman, E.P, Neau, D.B, Bezawada, P, Alasady, M.J, Varney, K.M, Toth, E.A, MacKerell Jr, A.D, Coop, A, Weber, D.J. | | Deposit date: | 2015-09-03 | | Release date: | 2016-01-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.742 Å) | | Cite: | Small Molecule Inhibitors of Ca(2+)-S100B Reveal Two Protein Conformations.
J.Med.Chem., 59, 2016
|
|
5DKN
 
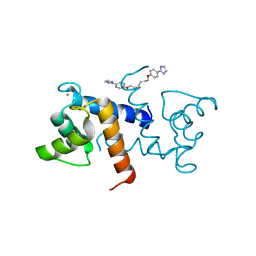 | | Crystal Structure of Calcium-loaded S100B bound to SBi4225 | | Descriptor: | 2,2'-[heptane-1,7-diylbis(oxybenzene-4,1-diyl)]bis(1H-imidazole), CALCIUM ION, Protein S100-B | | Authors: | Cavalier, M.C, Ansari, M.I, Pierce, A.D, Wilder, P.T, McKnight, L.E, Raman, E.P, Neau, D.B, Bezawada, P, Alasady, M.J, Varney, K.M, Toth, E.A, MacKerell Jr, A.D, Coop, A, Weber, D.J. | | Deposit date: | 2015-09-03 | | Release date: | 2016-01-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.528 Å) | | Cite: | Small Molecule Inhibitors of Ca(2+)-S100B Reveal Two Protein Conformations.
J.Med.Chem., 59, 2016
|
|
4FQO
 
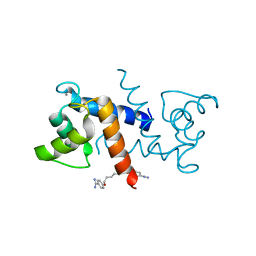 | | Crystal Structure of Calcium-Loaded S100B Bound to SBi4211 | | Descriptor: | 4,4'-[heptane-1,7-diylbis(oxy)]dibenzenecarboximidamide, CALCIUM ION, Protein S100-B | | Authors: | McKnight, L.E, Raman, E.P, Bezawada, P, Kudrimoti, S, Wilder, P.T, Hartman, K.G, Toth, E.A, Coop, A, MacKerrell, A.D, Weber, D.J. | | Deposit date: | 2012-06-25 | | Release date: | 2012-10-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure-Based Discovery of a Novel Pentamidine-Related Inhibitor of the Calcium-Binding Protein S100B.
ACS Med Chem Lett, 3, 2012
|
|
4DIR
 
 | |
4DK9
 
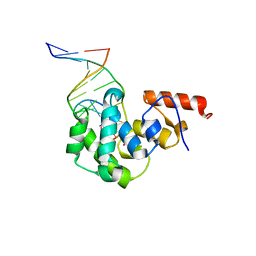 | | Crystal Structure of MBD4 Catalytic Domain Bound to Abasic DNA | | Descriptor: | 5'-D(*AP*AP*GP*AP*CP*GP*TP*GP*GP*AP*C)-3', 5'-D(*TP*GP*TP*CP*CP*AP*(3DR)P*GP*TP*CP*T)-3', Methyl-CpG-binding domain protein 4, ... | | Authors: | Manvilla, B.A, Toth, E.A, Drohat, A.C. | | Deposit date: | 2012-02-03 | | Release date: | 2012-04-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Crystal Structure of Human Methyl-Binding Domain IV Glycosylase Bound to Abasic DNA.
J.Mol.Biol., 420, 2012
|
|
1K1Q
 
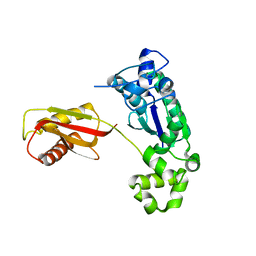 | | Crystal Structure of a DinB Family Error Prone DNA Polymerase from Sulfolobus solfataricus | | Descriptor: | DBH protein | | Authors: | Silvian, L.F, Toth, E.A, Pham, P, Goodman, M.F, Ellenberger, T. | | Deposit date: | 2001-09-25 | | Release date: | 2001-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of a DinB family error-prone DNA polymerase from Sulfolobus solfataricus.
Nat.Struct.Biol., 8, 2001
|
|
1K1S
 
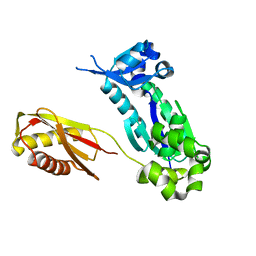 | | Crystal Structure of DinB from Sulfolobus solfataricus | | Descriptor: | DBH protein | | Authors: | Silvian, L.F, Toth, E.A, Pham, P, Goodman, M.F, Ellenberger, T. | | Deposit date: | 2001-09-25 | | Release date: | 2001-10-31 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of a DinB family error-prone DNA polymerase from Sulfolobus solfataricus.
Nat.Struct.Biol., 8, 2001
|
|
4KWV
 
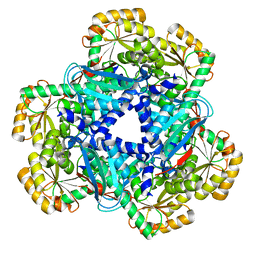 | | Crystal Structure of human apo-QPRT | | Descriptor: | Nicotinate-nucleotide pyrophosphorylase [carboxylating] | | Authors: | Malik, S.S, Patterson, D.N, Ncube, Z, Toth, E.A. | | Deposit date: | 2013-05-24 | | Release date: | 2013-10-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.797 Å) | | Cite: | The crystal structure of human quinolinic acid phosphoribosyltransferase in complex with its inhibitor phthalic acid.
Proteins, 82, 2014
|
|
4KWW
 
 | | The crystal structure of human quinolinic acid phosphoribosyltransferase in complex with its inhibitor phthalic acid | | Descriptor: | Nicotinate-nucleotide pyrophosphorylase [carboxylating], PHTHALIC ACID | | Authors: | Malik, S.S, Dimeka, P.N, Ncube, Z, Toth, E.A. | | Deposit date: | 2013-05-24 | | Release date: | 2013-10-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | The crystal structure of human quinolinic acid phosphoribosyltransferase in complex with its inhibitor phthalic acid.
Proteins, 82, 2014
|
|
4LND
 
 | | Crystal structure of human apurinic/apyrimidinic endonuclease 1 with essential Mg2+ cofactor | | Descriptor: | DNA-(apurinic or apyrimidinic site) lyase, MAGNESIUM ION | | Authors: | Manvilla, B.A, Pozharski, E, Toth, E.A, Drohat, A.C. | | Deposit date: | 2013-07-11 | | Release date: | 2013-11-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structure of human apurinic/apyrimidinic endonuclease 1 with the essential Mg(2+) cofactor.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
