2QCS
 
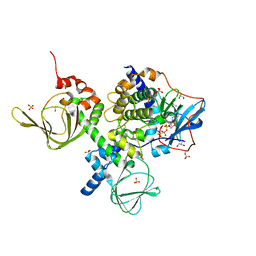 | | A complex structure between the Catalytic and Regulatory subunit of Protein Kinase A that represents the inhibited state | | Descriptor: | ACETATE ION, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Kim, C, Cheng, C.Y, Saldanha, A.S, Taylor, S.S. | | Deposit date: | 2007-06-19 | | Release date: | 2007-11-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | PKA-I holoenzyme structure reveals a mechanism for cAMP-dependent activation.
Cell(Cambridge,Mass.), 130, 2007
|
|
8GHS
 
 | | Empty HBV Cp183 capsid with importin-beta, subparticle reconstruction at 2-fold location | | Descriptor: | Capsid protein | | Authors: | Kim, C, Schlicksup, C.J, Hadden-Perilla, J.A, Wang, J.C.-Y, Zlotnick, A. | | Deposit date: | 2023-03-10 | | Release date: | 2023-08-09 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structure of the Hepatitis B virus capsid quasi-6-fold with a trapped C-terminal domain reveals capsid movements associated with domain exit.
J.Biol.Chem., 299, 2023
|
|
1KGZ
 
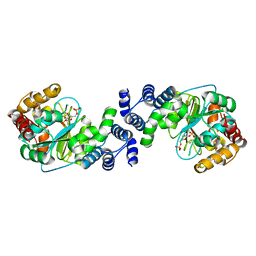 | | Crystal Structure Analysis of the Anthranilate Phosphoribosyltransferase from Erwinia carotovora (current name, Pectobacterium carotovorum) | | Descriptor: | 1-O-pyrophosphono-5-O-phosphono-alpha-D-ribofuranose, Anthranilate phosphoribosyltransferase, MANGANESE (II) ION, ... | | Authors: | Kim, C, Xuong, N.-H, Edwards, S, Madhusudan, Yee, M.-C, Spraggon, G, Mills, S.E. | | Deposit date: | 2001-11-28 | | Release date: | 2002-10-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Crystal Structure of Anthranilate
Phosphoribosyltransferase from the
Enterobacterium Pectobacterium carotovorum
FEBS Lett., 523, 2002
|
|
3FHI
 
 | |
1KHD
 
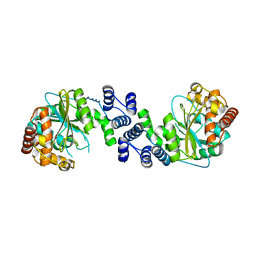 | | Crystal Structure Analysis of the anthranilate phosphoribosyltransferase from Erwinia carotovora at 1.9 resolution (current name, Pectobacterium carotovorum) | | Descriptor: | Anthranilate phosphoribosyltransferase | | Authors: | Kim, C, Xuong, N.-H, Edwards, S, Madhusudan, Yee, M.-C, Spraggon, G, Mills, S.E. | | Deposit date: | 2001-11-29 | | Release date: | 2002-10-23 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | The Crystal Structure of Anthranilate
Phosphoribosyltransferase from the
Enterobacterium Pectobacterium carotovorum
FEBS Lett., 523, 2002
|
|
6BQ8
 
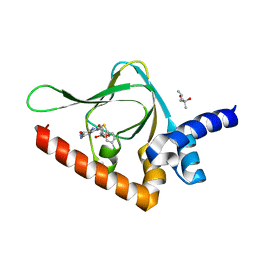 | | Joint X-ray/neutron structure of PKG II CNB-B domain in complex with 8-pCPT-cGMP | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-(~2~H_2_)amino-8-[(4-chlorophenyl)sulfanyl]-9-[(2S,4aR,6R,7R,7aS)-2-hydroxy-7-(~2~H)hydroxy-2-oxotetrahydro-2H,4H-2lambda~5~-furo[3,2-d][1,3,2]dioxaphosphinin-6-yl](~2~H)-1,9-dihydro-6H-purin-6-one, STRONTIUM ION, ... | | Authors: | Kim, C, Kovalevsky, A, Gerlits, O. | | Deposit date: | 2017-11-27 | | Release date: | 2018-03-21 | | Last modified: | 2024-04-03 | | Method: | NEUTRON DIFFRACTION (2 Å), X-RAY DIFFRACTION | | Cite: | Neutron Crystallography Detects Differences in Protein Dynamics: Structure of the PKG II Cyclic Nucleotide Binding Domain in Complex with an Activator.
Biochemistry, 57, 2018
|
|
8SSP
 
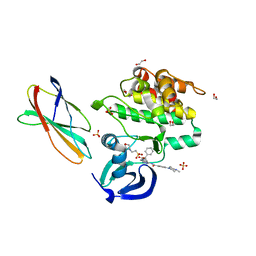 | | AurA bound to danusertib and activating monobody Mb1 | | Descriptor: | 1,2-ETHANEDIOL, 1,3-PROPANDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Ludewig, H, Kim, C, Kern, D. | | Deposit date: | 2023-05-08 | | Release date: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A biophysical framework for double-drugging kinases.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8SSO
 
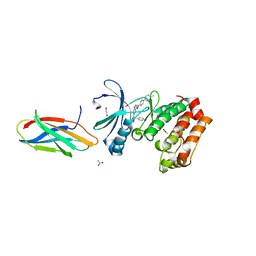 | | AurA bound to danusertib and inhibiting monobody Mb2 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Aurora kinase A, ... | | Authors: | Ludewig, H, Kim, C, Kern, D. | | Deposit date: | 2023-05-08 | | Release date: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | A biophysical framework for double-drugging kinases.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8SSN
 
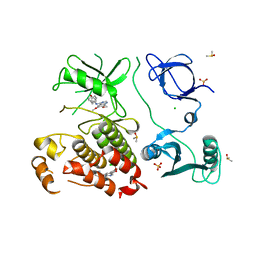 | | Abl kinase in complex with SKI and asciminib | | Descriptor: | 6,7-dimethoxy-N-(4-phenoxyphenyl)quinazolin-4-amine, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Ludewig, H, Kim, C, Kern, D. | | Deposit date: | 2023-05-08 | | Release date: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | A biophysical framework for double-drugging kinases.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
4QXK
 
 | | Joint X-ray/neutron structure of PKGIbeta in complex with cGMP | | Descriptor: | CYCLIC GUANOSINE MONOPHOSPHATE, SODIUM ION, cGMP-dependent protein kinase 1 | | Authors: | Kim, C, Gerlits, O, Kovalevsky, A, Huang, G.Y. | | Deposit date: | 2014-07-21 | | Release date: | 2014-11-12 | | Last modified: | 2024-02-28 | | Method: | NEUTRON DIFFRACTION (2.2 Å), X-RAY DIFFRACTION | | Cite: | Neutron Diffraction Reveals Hydrogen Bonds Critical for cGMP-Selective Activation: Insights for cGMP-Dependent Protein Kinase Agonist Design.
Biochemistry, 53, 2014
|
|
3NMD
 
 | | Crystal structure of the leucine zipper domain of cGMP dependent protein kinase I beta | | Descriptor: | GLYCEROL, HEXANE-1,6-DIOL, cGMP Dependent PRotein Kinase | | Authors: | Kim, C, Casteel, D.E, Smith-Nguyen, E.V, Sankaran, B, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2010-06-22 | | Release date: | 2010-09-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.272 Å) | | Cite: | A crystal structure of the cyclic GMP-dependent protein kinase I{beta} dimerization/docking domain reveals molecular details of isoform-specific anchoring.
J.Biol.Chem., 285, 2010
|
|
3FJQ
 
 | |
7PXZ
 
 | | Reduced form of SARS-CoV-2 Main Protease determined by XFEL radiation | | Descriptor: | 3C-like proteinase nsp5, CHLORIDE ION | | Authors: | Schubert, R, Reinke, P, Galchenkova, M, Oberthuer, D, Murillo, G.E.P, Kim, C, Bean, R, Turk, D, Hinrichs, W, Middendorf, P, Round, A, Schmidt, C, Mills, G, Kirkwood, H, Han, H, Koliyadu, J, Bielecki, J, Gelisio, L, Sikorski, M, Kloos, M, Vakilii, M, Yefanov, O.N, Vagovic, P, de-Wijn, R, Letrun, R, Guenther, S, White, T.A, Sato, T, Srinivasan, V, Kim, Y, Chretien, A, Han, S, Brognaro, H, Maracke, J, Knoska, J, Seychell, B.C, Brings, L, Norton-Baker, B, Geng, T, Dore, A.S, Uetrecht, C, Redecke, L, Beck, T, Lorenzen, K, Betzel, C, Mancuso, A.P, Bajt, S, Chapman, H.N, Meents, A, Lane, T.J. | | Deposit date: | 2021-10-08 | | Release date: | 2023-01-18 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | SARS-CoV-2 M pro responds to oxidation by forming disulfide and NOS/SONOS bonds.
Nat Commun, 15, 2024
|
|
7PZQ
 
 | | Oxidized form of SARS-CoV-2 Main Protease determined by XFEL radiation | | Descriptor: | 3C-like proteinase nsp5, DIMETHYL SULFOXIDE | | Authors: | Schubert, R, Reinke, P, Galchenkova, M, Oberthuer, D, Murillo, G.E.P, Kim, C, Bean, R, Turk, D, Hinrichs, W, Middendorf, P, Round, A, Schmidt, C, Mills, G, Kirkwood, H, Han, H, Koliyadu, J, Bielecki, J, Gelisio, L, Sikorski, M, Kloos, M, Vakilii, M, Yefanov, O.N, Vagovic, P, de-Wijn, R, Letrun, R, Guenther, S, White, T.A, Sato, T, Srinivasan, V, Kim, Y, Chretien, A, Han, S, Brognaro, H, Maracke, J, Knoska, J, Seychell, B.C, Brings, L, Norton-Baker, B, Geng, T, Dore, A.S, Uetrecht, C, Redecke, L, Beck, T, Lorenzen, K, Betzel, C, Mancuso, A.P, Bajt, S, Chapman, H.N, Meents, A, Lane, T.J. | | Deposit date: | 2021-10-13 | | Release date: | 2023-01-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | SARS-CoV-2 M pro responds to oxidation by forming disulfide and NOS/SONOS bonds.
Nat Commun, 15, 2024
|
|
6E49
 
 | | Pif1 peptide bound to PCNA trimer | | Descriptor: | ATP-dependent DNA helicase PIF1, Proliferating cell nuclear antigen | | Authors: | Buzovetsky, O, Kwon, Y, Pham, N.T, Kim, C, Ira, G, Sung, P, Xiong, Y. | | Deposit date: | 2018-07-17 | | Release date: | 2018-08-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Role of the Pif1-PCNA Complex in Pol delta-Dependent Strand Displacement DNA Synthesis and Break-Induced Replication.
Cell Rep, 21, 2017
|
|
4DBM
 
 | | Aplysia californica-AChBP in complex with triazole 18 | | Descriptor: | (3-exo)-8,8-dimethyl-3-(4-{[(1-methyl-2-oxo-1,2-dihydroquinolin-4-yl)oxy]methyl}-1H-1,2,3-triazol-1-yl)-8-azoniabicyclo[3.2.1]octane, 2-acetamido-2-deoxy-beta-D-glucopyranose, Soluble acetylcholine receptor, ... | | Authors: | Nemecz, A, Yamauchi, J.G, Kim, C. | | Deposit date: | 2012-01-16 | | Release date: | 2012-03-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Generation of candidate ligands for nicotinic acetylcholine receptors via in situ click chemistry with a soluble acetylcholine binding protein template.
J.Am.Chem.Soc., 134, 2012
|
|
5KJY
 
 | | Co-crystal structure of PKA RI alpha CNB-B mutant (G316R/A336T) with cAMP | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, cAMP-dependent protein kinase type I-alpha regulatory subunit | | Authors: | Lorenz, R, Moon, E, Kim, J.J, Huang, G.Y, Kim, C, Herberg, F.W. | | Deposit date: | 2016-06-20 | | Release date: | 2017-06-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mutations of PKA cyclic nucleotide-binding domains reveal novel aspects of cyclic nucleotide selectivity.
Biochem. J., 474, 2017
|
|
5KJX
 
 | | Co-crystal Structure of PKA RI alpha CNB-B domain with cAMP | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, cAMP-dependent protein kinase type I-alpha regulatory subunit | | Authors: | Lorenz, R, Moon, E, Kim, J.J, Huang, G.Y, Kim, C, Herberg, F.W. | | Deposit date: | 2016-06-20 | | Release date: | 2017-06-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mutations of PKA cyclic nucleotide-binding domains reveal novel aspects of cyclic nucleotide selectivity.
Biochem. J., 474, 2017
|
|
5KJZ
 
 | | Co-crystal structure of PKA RI alpha CNB-B mutant (G316R/A336T) with cGMP | | Descriptor: | CYCLIC GUANOSINE MONOPHOSPHATE, GLYCEROL, cAMP-dependent protein kinase type I-alpha regulatory subunit | | Authors: | Lorenz, R, Moon, E, Kim, J.J, Huang, G.Y, Kim, C, Herberg, F.W. | | Deposit date: | 2016-06-20 | | Release date: | 2017-06-28 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.347 Å) | | Cite: | Mutations of PKA cyclic nucleotide-binding domains reveal novel aspects of cyclic nucleotide selectivity.
Biochem. J., 474, 2017
|
|
4Z07
 
 | | Co-crystal structure of the tandem CNB (CNB-A/B) domains of human PKG I beta with cGMP | | Descriptor: | CYCLIC GUANOSINE MONOPHOSPHATE, ISOPROPYL ALCOHOL, SULFATE ION, ... | | Authors: | Kim, J.J, Reger, A.S, Arold, S.T, Kim, C. | | Deposit date: | 2015-03-25 | | Release date: | 2016-04-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of PKG I:cGMP Complex Reveals a cGMP-Mediated Dimeric Interface that Facilitates cGMP-Induced Activation.
Structure, 24, 2016
|
|
4KU8
 
 | | Structures of PKGI Reveal a cGMP-Selective Activation Mechanism | | Descriptor: | GLYCINE, cGMP-dependent Protein Kinase 1 | | Authors: | Huang, G.Y, Kim, J.J, Reger, A.S, Lorenz, R, Moon, E.W, Casteel, D.E, Sankaran, B, Herberg, F.W, Kim, C. | | Deposit date: | 2013-05-21 | | Release date: | 2014-01-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.994 Å) | | Cite: | Structural Basis for Cyclic-Nucleotide Selectivity and cGMP-Selective Activation of PKG I.
Structure, 22, 2014
|
|
4KU7
 
 | | Structures of PKGI Reveal a cGMP-Selective Activation Mechanism | | Descriptor: | CYCLIC GUANOSINE MONOPHOSPHATE, IODIDE ION, cGMP-dependent protein kinase 1 | | Authors: | Huang, G.Y, Kim, J.J, Reger, A.S, Lorenz, R, Moon, E.W, Casteel, D.E, Sankaran, B, Herberg, F.W, Kim, C. | | Deposit date: | 2013-05-21 | | Release date: | 2014-01-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural Basis for Cyclic-Nucleotide Selectivity and cGMP-Selective Activation of PKG I.
Structure, 22, 2014
|
|
6VTX
 
 | | Crystal structure of human KLF4 zinc finger DNA binding domain in complex with NANOG DNA | | Descriptor: | DNA (5'-D(*AP*GP*GP*GP*GP*GP*TP*GP*TP*GP*CP*C)-3'), DNA (5'-D(*GP*GP*CP*AP*CP*AP*CP*CP*CP*CP*CP*T)-3'), Krueppel-like factor 4, ... | | Authors: | Sharma, R, Sharma, S, Choi, K.J, Ferreon, A.C.M, Ferreon, J.C, Sankaran, B, MacKenzie, K.R, Kim, C. | | Deposit date: | 2020-02-13 | | Release date: | 2021-09-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Liquid condensation of reprogramming factor KLF4 with DNA provides a mechanism for chromatin organization.
Nat Commun, 12, 2021
|
|
7KN0
 
 | | Structure of the integrin aIIb(W968V)b3 transmembrane complex | | Descriptor: | Integrin alpha-IIb, Integrin beta-3 | | Authors: | Situ, A.J, Kim, J, An, W, Kim, C, Ulmer, T.S. | | Deposit date: | 2020-11-03 | | Release date: | 2021-09-15 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Insight Into Pathological Integrin alpha IIb beta 3 Activation From Safeguarding The Inactive State.
J.Mol.Biol., 433, 2021
|
|
8VF6
 
 | | Crystal structure of Serine/threonine-protein kinase 33 (STK33) Kinase Domain in complex with inhibitor CDD-2211 | | Descriptor: | Serine/threonine-protein kinase 33, {3-[([1,1'-biphenyl]-2-yl)ethynyl]-1H-indazol-5-yl}[(3R)-3-(dimethylamino)pyrrolidin-1-yl]methanone | | Authors: | Ta, H.M, Kim, C, Ku, K.A, Matzuk, M.M. | | Deposit date: | 2023-12-21 | | Release date: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Reversible male contraception by targeted inhibition of serine/threonine kinase 33.
Science, 384, 2024
|
|
